The circadian clock and darkness control natural competence in cyanobacteria
- PMID: 32245943
- PMCID: PMC7125226
- DOI: 10.1038/s41467-020-15384-9
The circadian clock and darkness control natural competence in cyanobacteria
Abstract
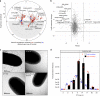
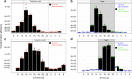
The cyanobacterium Synechococcus elongatus is a model organism for the study of circadian rhythms. It is naturally competent for transformation-that is, it takes up DNA from the environment, but the underlying mechanisms are unclear. Here, we use a genome-wide screen to identify genes required for natural transformation in S. elongatus, including genes encoding a conserved Type IV pilus, genes known to be associated with competence in other bacteria, and others. Pilus biogenesis occurs daily in the morning, while natural transformation is maximal when the onset of darkness coincides with the dusk circadian peak. Thus, the competence state in cyanobacteria is regulated by the circadian clock and can adapt to seasonal changes of day length.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

