A harmonized meta-knowledgebase of clinical interpretations of somatic genomic variants in cancer
- PMID: 32246132
- PMCID: PMC7127986
- DOI: 10.1038/s41588-020-0603-8
A harmonized meta-knowledgebase of clinical interpretations of somatic genomic variants in cancer
Abstract
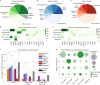
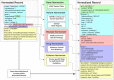
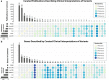
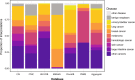
Precision oncology relies on accurate discovery and interpretation of genomic variants, enabling individualized diagnosis, prognosis and therapy selection. We found that six prominent somatic cancer variant knowledgebases were highly disparate in content, structure and supporting primary literature, impeding consensus when evaluating variants and their relevance in a clinical setting. We developed a framework for harmonizing variant interpretations to produce a meta-knowledgebase of 12,856 aggregate interpretations. We demonstrated large gains in overlap between resources across variants, diseases and drugs as a result of this harmonization. We subsequently demonstrated improved matching between a patient cohort and harmonized interpretations of potential clinical significance, observing an increase from an average of 33% per individual knowledgebase to 57% in aggregate. Our analyses illuminate the need for open, interoperable sharing of variant interpretation data. We also provide a freely available web interface (search.cancervariants.org) for exploring the harmonized interpretations from these six knowledgebases.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Grants and funding
- K99 HG010157/HG/NHGRI NIH HHS/United States
- U41 HG006834/HG/NHGRI NIH HHS/United States
- U01 CA209936/CA/NCI NIH HHS/United States
- U01 CA231844/CA/NCI NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- R24 OD011883/OD/NIH HHS/United States
- F32 CA206247/CA/NCI NIH HHS/United States
- MRC_/Medical Research Council/United Kingdom
- R00 HG007940/HG/NHGRI NIH HHS/United States
- U41 HG009649/HG/NHGRI NIH HHS/United States
- CRUK_/Cancer Research UK/United Kingdom
- R01 CA180778/CA/NCI NIH HHS/United States
- U24 CA237719/CA/NCI NIH HHS/United States
- U54 HG007990/HG/NHGRI NIH HHS/United States
- U41 HG009650/HG/NHGRI NIH HHS/United States
- U01 CA217862/CA/NCI NIH HHS/United States
- U01 HG007437/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

