Cell Type-Specific Intralocus Interactions Reveal Oligodendrocyte Mechanisms in MS
- PMID: 32246942
- PMCID: PMC7426147
- DOI: 10.1016/j.cell.2020.03.002
Cell Type-Specific Intralocus Interactions Reveal Oligodendrocyte Mechanisms in MS
Abstract
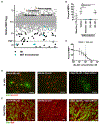
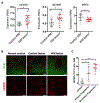
Multiple sclerosis (MS) is an autoimmune disease characterized by attack on oligodendrocytes within the central nervous system (CNS). Despite widespread use of immunomodulatory therapies, patients may still face progressive disability because of failure of myelin regeneration and loss of neurons, suggesting additional cellular pathologies. Here, we describe a general approach for identifying specific cell types in which a disease allele exerts a pathogenic effect. Applying this approach to MS risk loci, we pinpoint likely pathogenic cell types for 70%. In addition to T cell loci, we unexpectedly identified myeloid- and CNS-specific risk loci, including two sites that dysregulate transcriptional pause release in oligodendrocytes. Functional studies demonstrated inhibition of transcriptional elongation is a dominant pathway blocking oligodendrocyte maturation. Furthermore, pause release factors are frequently dysregulated in MS brain tissue. These data implicate cell-intrinsic aberrations outside of the immune system and suggest new avenues for therapeutic development. VIDEO ABSTRACT.
Keywords: GWAS; cell type; epigenomics; genetic risk; multiple sclerosis; oligodendrocytes; outside variants; population genetics; remyelination; transcriptional pause release.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests P.J.T. and D.J.A are co-founders and consultants for Convelo Therapeutics, which has licensed patents unrelated to the current study. P.J.T., D.J.A., and Case Western Reserve University retain equity in Convelo Therapeutics. D.C.F. is currently an employee and shareholder of Convelo Therapeutics.
Figures



References
-
- Adams D, Altucci L, Antonarakis SE, Ballesteros J, Beck S, Bird A, Bock C, Boehm B, Campo E, Caricasole A, et al. (2012). BLUEPRINT to decode the epigenetic signature written in blood. Nat. Biotechnol. 30, 224–226. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

