New strategies for profiling and characterization of human milk oligosaccharides
- PMID: 32248230
- PMCID: PMC7526734
- DOI: 10.1093/glycob/cwaa028
New strategies for profiling and characterization of human milk oligosaccharides
Abstract
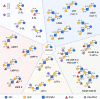
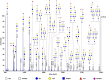
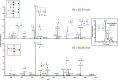
Human breast milk is an incredibly rich and complex biofluid composed of proteins, lipids and complex carbohydrates, including a diverse repertoire of free human milk oligosaccharides (HMOs). Strikingly, HMOs are not digested by the infant but function as prebiotics for bacterial strains associated with numerous benefits. Considering the broad variety of beneficial effects of HMOs, and the vast number of factors that affect breast milk composition, the analysis of HMO diversity and complexity is of utmost relevance. Using human milk samples from a cohort of Bangladeshi mothers participating in a study on malnutrition and stunting in children, we have characterized breast milk oligosaccharide composition by means of permethylation followed by liquid chromatography coupled with high-resolution tandem mass spectrometry (LC-MS/MS) analysis. This approach identified over 100 different glycoforms and showed a wide diversity of milk composition, with a predominance of fucosylated and sialylated HMOs over nonmodified HMOs. We observed that these samples contain on average 80 HMOs, with the highest permethylated masses detected being >5000 mass units. Here we report an easily implemented method developed for the separation, characterization and relative quantitation of large arrays of HMOs, including higher molecular weight sialylated HMOs. Our ultimate goal is to create a simple, high-throughput method, which can be used for full characterization of sialylated and/or fucosylated HMOs. These results demonstrate how current analytical techniques can be applied to characterize human milk composition, providing new tools to help the scientific community shed new light on the impact of HMOs during infant development.
Keywords: CID; LC-NSI-MS/MS; MALDI-TOF-MS; human milk oligosaccharides (HMOs); structural analysis.
© The Author(s) 2020. Published by Oxford University Press.
Figures



Similar articles
-
Robust and High-Resolution All-Ion Fragmentation LC-ESI-IM-MS Analysis for In-Depth Characterization or Profiling of Up to 200 Human Milk Oligosaccharides.Anal Chem. 2025 Mar 18;97(10):5563-5574. doi: 10.1021/acs.analchem.4c06081. Epub 2025 Mar 6. Anal Chem. 2025. PMID: 40047520 Free PMC article.
-
Comparative analysis of native and permethylated human milk oligosaccharides by liquid chromatography coupled to high resolution mass spectrometry.J Chromatogr B Analyt Technol Biomed Life Sci. 2017 Dec 15;1071:49-57. doi: 10.1016/j.jchromb.2017.03.028. Epub 2017 Mar 27. J Chromatogr B Analyt Technol Biomed Life Sci. 2017. PMID: 28377082
-
Label-free targeted LC-ESI-MS2 analysis of human milk oligosaccharides (HMOS) and related human milk groups with enhanced structural selectivity.Anal Bioanal Chem. 2019 Jan;411(1):231-250. doi: 10.1007/s00216-018-1434-7. Epub 2018 Nov 16. Anal Bioanal Chem. 2019. PMID: 30443773
-
Analytical characterization of human milk oligosaccharides - potential applications in pharmaceutical analysis.J Pharm Biomed Anal. 2017 Nov 30;146:168-178. doi: 10.1016/j.jpba.2017.08.039. Epub 2017 Aug 31. J Pharm Biomed Anal. 2017. PMID: 28881314 Review.
-
Recent advances on separation and characterization of human milk oligosaccharides.Electrophoresis. 2016 Jun;37(11):1514-24. doi: 10.1002/elps.201500477. Epub 2016 Feb 24. Electrophoresis. 2016. PMID: 26801168 Review.
Cited by
-
Development of a cyclic ion mobility spectrometry-mass spectrometry-based collision cross-section database of permethylated human milk oligosaccharides.J Mass Spectrom. 2024 Aug;59(8):e5076. doi: 10.1002/jms.5076. J Mass Spectrom. 2024. PMID: 39041358 Free PMC article.
-
Comparative analysis of oligosaccharides in the milk of human and animals by using LC-QE-HF-MS.Food Chem X. 2023 May 9;18:100705. doi: 10.1016/j.fochx.2023.100705. eCollection 2023 Jun 30. Food Chem X. 2023. PMID: 37397214 Free PMC article.
-
Recent advances in the science of human milk oligosaccharides.BBA Adv. 2025 Jan 24;7:100136. doi: 10.1016/j.bbadva.2024.100136. eCollection 2025. BBA Adv. 2025. PMID: 39991261 Free PMC article.
-
Robust and High-Resolution All-Ion Fragmentation LC-ESI-IM-MS Analysis for In-Depth Characterization or Profiling of Up to 200 Human Milk Oligosaccharides.Anal Chem. 2025 Mar 18;97(10):5563-5574. doi: 10.1021/acs.analchem.4c06081. Epub 2025 Mar 6. Anal Chem. 2025. PMID: 40047520 Free PMC article.
-
Human milk oligosaccharides: bridging the gap in intestinal microbiota between mothers and infants.Front Cell Infect Microbiol. 2025 Jan 6;14:1386421. doi: 10.3389/fcimb.2024.1386421. eCollection 2024. Front Cell Infect Microbiol. 2025. PMID: 39835278 Free PMC article. Review.
References
-
- Apte A, Meitei NS. 2010. Bioinformatics in glycomics: Glycan characterization with mass spectrometric data using SimGlycan. Methods Mol Biol. 600:269–281. - PubMed
-
- Asakuma S, Urashima T, Akahori M, Obayashi H, Nakamura T, Kimura K, Watanabe Y, Arai I, Sanai Y. 2008. Variation of major neutral oligosaccharides levels in human colostrum. Eur J Clin Nutr. 62:488–494. - PubMed
-
- Ashline DJ, Zhang H, Reinhold VN. 2017. Isomeric complexity of glycosylation documented by MS(n). Anal Bioanal Chem. 409:439–451. - PubMed

