SUMOylation of the transcription factor ZFHX3 at Lys-2806 requires SAE1, UBC9, and PIAS2 and enhances its stability and function in cell proliferation
- PMID: 32249212
- PMCID: PMC7212658
- DOI: 10.1074/jbc.RA119.012338
SUMOylation of the transcription factor ZFHX3 at Lys-2806 requires SAE1, UBC9, and PIAS2 and enhances its stability and function in cell proliferation
Abstract
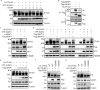
SUMOylation is a posttranslational modification (PTM) at a lysine residue and is crucial for the proper functions of many proteins, particularly of transcription factors, in various biological processes. Zinc finger homeobox 3 (ZFHX3), also known as AT motif-binding factor 1 (ATBF1), is a large transcription factor that is active in multiple pathological processes, including atrial fibrillation and carcinogenesis, and in circadian regulation and development. We have previously demonstrated that ZFHX3 is SUMOylated at three or more lysine residues. Here, we investigated which enzymes regulate ZFHX3 SUMOylation and whether SUMOylation modulates ZFHX3 stability and function. We found that SUMO1, SUMO2, and SUMO3 each are conjugated to ZFHX3. Multiple lysine residues in ZFHX3 were SUMOylated, but Lys-2806 was the major SUMOylation site, and we also found that it is highly conserved among ZFHX3 orthologs from different animal species. Using molecular analyses, we identified the enzymes that mediate ZFHX3 SUMOylation; these included SUMO1-activating enzyme subunit 1 (SAE1), an E1-activating enzyme; SUMO-conjugating enzyme UBC9 (UBC9), an E2-conjugating enzyme; and protein inhibitor of activated STAT2 (PIAS2), an E3 ligase. Multiple analyses established that both SUMO-specific peptidase 1 (SENP1) and SENP2 deSUMOylate ZFHX3. SUMOylation at Lys-2806 enhanced ZFHX3 stability by interfering with its ubiquitination and proteasomal degradation. Functionally, Lys-2806 SUMOylation enabled ZFHX3-mediated cell proliferation and xenograft tumor growth of the MDA-MB-231 breast cancer cell line. These findings reveal the enzymes involved in, and the functional consequences of, ZFHX3 SUMOylation, insights that may help shed light on ZFHX3's roles in various cellular and pathophysiological processes.
Keywords: SUMO-conjugating enzyme UBC9; SUMO-specific peptidase 1 (SENP1); SUMO1-activating enzyme subunit 1 (SAE1); SUMOylation; UBC9; cancer; cell proliferation; posttranslational modification (PTM); proliferation; protein inhibitor of activated STAT2 (PIAS2); protein stability; transcription factor; zinc finger homeobox 3 (ZFHX3).
© 2020 Wu et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




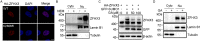

Similar articles
-
Identification of a non-covalent ternary complex formed by PIAS1, SUMO1, and UBC9 proteins involved in transcriptional regulation.J Biol Chem. 2013 Dec 20;288(51):36312-27. doi: 10.1074/jbc.M113.486845. Epub 2013 Oct 30. J Biol Chem. 2013. PMID: 24174529 Free PMC article.
-
Structural insights into the regulation of the human E2∼SUMO conjugate through analysis of its stable mimetic.J Biol Chem. 2023 Jul;299(7):104870. doi: 10.1016/j.jbc.2023.104870. Epub 2023 May 27. J Biol Chem. 2023. PMID: 37247759 Free PMC article.
-
Akt SUMOylation regulates cell proliferation and tumorigenesis.Cancer Res. 2013 Sep 15;73(18):5742-53. doi: 10.1158/0008-5472.CAN-13-0538. Epub 2013 Jul 24. Cancer Res. 2013. PMID: 23884910
-
SUMO pathway components as possible cancer biomarkers.Future Oncol. 2015;11(11):1599-610. doi: 10.2217/fon.15.41. Future Oncol. 2015. PMID: 26043214 Review.
-
Mechanisms and functions of SUMOylation in health and disease: a review focusing on immune cells.J Biomed Sci. 2024 Jan 27;31(1):16. doi: 10.1186/s12929-024-01003-y. J Biomed Sci. 2024. PMID: 38280996 Free PMC article. Review.
Cited by
-
SENP1 modulates microglia-mediated neuroinflammation toward intermittent hypoxia-induced cognitive decline through the de-SUMOylation of NEMO.J Cell Mol Med. 2021 Jul;25(14):6841-6854. doi: 10.1111/jcmm.16689. Epub 2021 Jun 13. J Cell Mol Med. 2021. PMID: 34120412 Free PMC article.
-
The SUMOylation and ubiquitination crosstalk in cancer.J Cancer Res Clin Oncol. 2023 Nov;149(17):16123-16146. doi: 10.1007/s00432-023-05310-z. Epub 2023 Aug 28. J Cancer Res Clin Oncol. 2023. PMID: 37640846 Free PMC article. Review.
-
SENP1 mediates zinc-induced ZnT6 deSUMOylation at Lys-409 involved in the regulation of zinc metabolism in Golgi apparatus.Cell Mol Life Sci. 2024 Oct 5;81(1):422. doi: 10.1007/s00018-024-05452-4. Cell Mol Life Sci. 2024. PMID: 39367979 Free PMC article.
-
Integrative bioinformatics analysis reveals STAT2 as a novel biomarker of inflammation-related cardiac dysfunction in atrial fibrillation.Open Med (Wars). 2023 Nov 9;18(1):20230834. doi: 10.1515/med-2023-0834. eCollection 2023. Open Med (Wars). 2023. PMID: 38025532 Free PMC article.
-
ATBF1 is a potential diagnostic marker of histological grade and functions via WNT5A in breast cancer.BMC Cancer. 2022 Dec 7;22(1):1280. doi: 10.1186/s12885-022-10380-2. BMC Cancer. 2022. PMID: 36476423 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

