Quantification and Phylogenetic Analysis of Ammonia Oxidizers on Biofilm Carriers in a Full-Scale Wastewater Treatment Plant
- PMID: 32249239
- PMCID: PMC7308565
- DOI: 10.1264/jsme2.ME19140
Quantification and Phylogenetic Analysis of Ammonia Oxidizers on Biofilm Carriers in a Full-Scale Wastewater Treatment Plant
Abstract
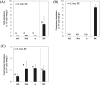
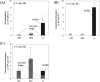
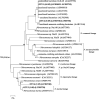
Biofilm carriers have been used to remove ammonia in several wastewater treatment plants (WWTPs) in Japan. However, the abundance and species of ammonia oxidizers in the biofilms formed on the surface of carriers in full-scale operational WWTP tanks remain unclear. In the present study, we conducted quantitative PCR and PCR cloning of the amoA genes of ammonia-oxidizing bacteria and archaea (AOB and AOA) and a complete ammonia oxidizer (comammox) in the biofilm formed on the carriers in a full-scale WWTP. The quantification of amoA genes showed that the abundance of AOB and comammox was markedly greater in the biofilm than in the activated sludge suspended in a tank solution of the WWTP, while AOA was not detected in the biofilm or the activated sludge. A phylogenetic analysis of amoA genes revealed that as-yet-uncultivated comammox Nitrospira and uncultured AOB Nitrosomonas were predominant in the biofilm. The present results suggest that the biofilm formed on the surface of carriers enable comammox Nitrospira and AOB Nitrosomonas to co-exist and remain in the full-scale WWTP tank surveyed in this study.
Keywords: WWTP; ammonia-oxidizing bacteria; biofilm; comammox; real-time PCR.
Figures




Similar articles
-
The more important role of archaea than bacteria in nitrification of wastewater treatment plants in cold season despite their numerical relationships.Water Res. 2018 Nov 15;145:552-561. doi: 10.1016/j.watres.2018.08.066. Epub 2018 Aug 30. Water Res. 2018. PMID: 30199800
-
Comammox Nitrospira among dominant ammonia oxidizers within aquarium biofilter microbial communities.Appl Environ Microbiol. 2024 Jul 24;90(7):e0010424. doi: 10.1128/aem.00104-24. Epub 2024 Jun 20. Appl Environ Microbiol. 2024. PMID: 38899882 Free PMC article.
-
Identification and quantification of bacteria and archaea responsible for ammonia oxidation in different activated sludge of full-scale wastewater treatment plants.J Environ Sci Health A Tox Hazard Subst Environ Eng. 2015;50(2):169-75. doi: 10.1080/10934529.2014.975535. J Environ Sci Health A Tox Hazard Subst Environ Eng. 2015. PMID: 25560263
-
Nitrospira as versatile nitrifiers: Taxonomy, ecophysiology, genome characteristics, growth, and metabolic diversity.J Basic Microbiol. 2021 Feb;61(2):88-109. doi: 10.1002/jobm.202000485. Epub 2021 Jan 14. J Basic Microbiol. 2021. PMID: 33448079 Review.
-
Ammonia-oxidizing archaea and complete ammonia-oxidizing Nitrospira in water treatment systems.Water Res X. 2022 Mar 14;15:100131. doi: 10.1016/j.wroa.2022.100131. eCollection 2022 May 1. Water Res X. 2022. PMID: 35402889 Free PMC article. Review.
Cited by
-
Distributions, interactions, and dynamics of prokaryotes and phages in a hybrid biological wastewater treatment system.Microbiome. 2024 Jul 22;12(1):134. doi: 10.1186/s40168-024-01853-6. Microbiome. 2024. PMID: 39039555 Free PMC article.
References
-
- Almstrand R., Lydmark P., Sörensson F., and Hermansson M. (2011) Nitrification potential and population dynamics of nitrifying bacterial biofilms in response to controlled shifts of ammonium concentrations in wastewater trickling filters. Bioresour Technol 102: 7685–7691. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

