Organoid cultures from normal and cancer-prone human breast tissues preserve complex epithelial lineages
- PMID: 32249764
- PMCID: PMC7136203
- DOI: 10.1038/s41467-020-15548-7
Organoid cultures from normal and cancer-prone human breast tissues preserve complex epithelial lineages
Abstract
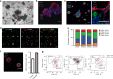
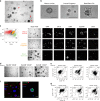
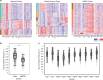
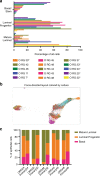
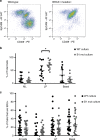
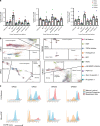
Recently, organoid technology has been used to generate a large repository of breast cancer organoids. Here we present an extensive evaluation of the ability of organoid culture technology to preserve complex stem/progenitor and differentiated cell types via long-term propagation of normal human mammary tissues. Basal/stem and luminal progenitor cells can differentiate in culture to generate mature basal and luminal cell types, including ER+ cells that have been challenging to maintain in culture. Cells associated with increased cancer risk can also be propagated. Single-cell analyses of matched organoid cultures and native tissues by mass cytometry for 38 markers provide a higher resolution representation of the multiple mammary epithelial cell types in the organoids, and demonstrate that protein expression patterns of the tissue of origin can be preserved in culture. These studies indicate that organoid cultures provide a valuable platform for studies of mammary differentiation, transformation, and breast cancer risk.
Conflict of interest statement
N.S. and H.C. declare that they are inventors on several patents related to organoid technology.
Figures


References
-
- Stampfer MR, Yaswen P. Culture systems for study of human mammary epithelial cell proliferation, differentiation and transformation. Cancer Surv. 1993;18:7–34. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

