Dynamic genome evolution and complex virocell metabolism of globally-distributed giant viruses
- PMID: 32249765
- PMCID: PMC7136201
- DOI: 10.1038/s41467-020-15507-2
Dynamic genome evolution and complex virocell metabolism of globally-distributed giant viruses
Abstract
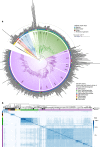
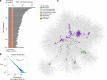
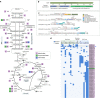
The discovery of eukaryotic giant viruses has transformed our understanding of the limits of viral complexity, but the extent of their encoded metabolic diversity remains unclear. Here we generate 501 metagenome-assembled genomes of Nucleo-Cytoplasmic Large DNA Viruses (NCLDV) from environments around the globe, and analyze their encoded functional capacity. We report a remarkable diversity of metabolic genes in widespread giant viruses, including many involved in nutrient uptake, light harvesting, and nitrogen metabolism. Surprisingly, numerous NCLDV encode the components of glycolysis and the TCA cycle, suggesting that they can re-program fundamental aspects of their host's central carbon metabolism. Our phylogenetic analysis of NCLDV metabolic genes and their cellular homologs reveals distinct clustering of viral sequences into divergent clades, indicating that these genes are virus-specific and were acquired in the distant past. Overall our findings reveal that giant viruses encode complex metabolic capabilities with evolutionary histories largely independent of cellular life, strongly implicating them as important drivers of global biogeochemical cycles.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

