Massive analysis of 64,628 bacterial genomes to decipher water reservoir and origin of mobile colistin resistance genes: is there another role for these enzymes?
- PMID: 32249837
- PMCID: PMC7136264
- DOI: 10.1038/s41598-020-63167-5
Massive analysis of 64,628 bacterial genomes to decipher water reservoir and origin of mobile colistin resistance genes: is there another role for these enzymes?
Abstract
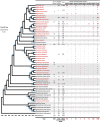
Since 2015, new worrying colistin resistance mechanism, mediated by mcr-1 gene has been reported worldwide along with eight newly described variants but their source(s) and reservoir(s) remain largely unexplored. Here, we conducted a massive bioinformatic analysis of bacterial genomes to investigate the reservoir and origin of mcr variants. We identified 13'658 MCR-1 homologous sequences in 494 bacterial genera. Moreover, analysis of 64'628 bacterial genomes (60 bacterial genera and 1'047 species) allows identifying a total of 6'651 significant positive hits (coverage >90% and similarity >50%) with the nine MCR variants from 39 bacterial genera and more than 1'050 species. A high number of MCR-1 was identified in Escherichia coli (n = 862). Interestingly, while almost all variants were identified in bacteria from different sources (i.e. human, animal, and environment), the last variant, MCR-9, was exclusively detected in bacteria from human. Although these variants could be identified in bacteria from human and animal sources, we found plenty MCR variants in unsuspected bacteria from environmental origin, especially from water sources. The ubiquitous presence of mcr variants in bacteria from water likely suggests another role in the biosphere of these enzymes as an unknown defense system against natural antimicrobial peptides and/or bacteriophage predation.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

