The pan-cancer landscape of netrin family reveals potential oncogenic biomarkers
- PMID: 32251318
- PMCID: PMC7090012
- DOI: 10.1038/s41598-020-62117-5
The pan-cancer landscape of netrin family reveals potential oncogenic biomarkers
Abstract
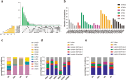
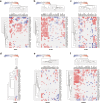
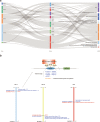
Recent cancer studies have found that the netrin family of proteins plays vital roles in the development of some cancers. However, the functions of the many variants of these proteins in cancer remain incompletely understood. In this work, we used the most comprehensive database available, including more than 10000 samples across more than 30 tumor types, to analyze the six members of the netrin family. We performed comprehensive analysis of genetic change and expression of the netrin genes and analyzed epigenetic and pathway relationships, as well as the correlation of expression of these proteins with drug sensitivity. Although the mutation rate of the netrin family is low in pan-cancer, among the tumor patients with netrin mutations, the highest number are Uterine Corpus Endometrial Carcinoma patients, accounting for 13.6% of cases (54 of 397). Interestingly, the highest mutation rate of a netrin family member is 38% for NTNG1 (152 of 397). Netrin proteins may participate in the development of endocrine-related tumors and sex hormone-targeting organ tumors. Additionally, the participation of NTNG1 and NTNG2 in various cancers shows their potential for use as new tumor markers and therapeutic targets. This analysis provides a broad molecular perspective of this protein family and suggests some new directions for the treatment of cancer.
Conflict of interest statement
The authors declare no competing interests.
Figures






Similar articles
-
Netrin G1: its downregulation in the nucleus accumbens of cocaine-conditioned mice and genetic association in human cocaine dependence.Addict Biol. 2018 Jan;23(1):448-460. doi: 10.1111/adb.12485. Epub 2017 Jan 11. Addict Biol. 2018. PMID: 28074533
-
Netrin-G2 dysfunction causes a Rett-like phenotype with areflexia.Hum Mutat. 2020 Feb;41(2):476-486. doi: 10.1002/humu.23945. Epub 2019 Nov 15. Hum Mutat. 2020. PMID: 31692205
-
Decreased mRNA expression of netrin-G1 and netrin-G2 in the temporal lobe in schizophrenia and bipolar disorder.Neuropsychopharmacology. 2008 Mar;33(4):933-45. doi: 10.1038/sj.npp.1301457. Epub 2007 May 16. Neuropsychopharmacology. 2008. PMID: 17507910 Free PMC article.
-
Netrin-1 in cancer: Potential biomarker and therapeutic target?Tumour Biol. 2017 Apr;39(4):1010428317698388. doi: 10.1177/1010428317698388. Tumour Biol. 2017. PMID: 28443497 Review.
-
Netrin-1: A regulator of cancer cell motility?Eur J Cell Biol. 2016 Nov;95(11):513-520. doi: 10.1016/j.ejcb.2016.10.002. Epub 2016 Oct 19. Eur J Cell Biol. 2016. PMID: 27793362 Review.
Cited by
-
A risk model based on lncRNA-miRNA-mRNA gene signature for predicting prognosis of patients with bladder cancer.Cancer Biomark. 2024;39(4):277-287. doi: 10.3233/CBM-230216. Cancer Biomark. 2024. PMID: 38306023 Free PMC article.
-
"Netrin-slaying nanoparticles: A new frontier in cancer therapy".BBA Adv. 2025 Jun 10;8:100163. doi: 10.1016/j.bbadva.2025.100163. eCollection 2025. BBA Adv. 2025. PMID: 40584519 Free PMC article. Review.
-
PharmacoDB 2.0: improving scalability and transparency of in vitro pharmacogenomics analysis.Nucleic Acids Res. 2022 Jan 7;50(D1):D1348-D1357. doi: 10.1093/nar/gkab1084. Nucleic Acids Res. 2022. PMID: 34850112 Free PMC article.
-
Hepatic Expression of NTN4 and Its Receptors in Patients with Hepatocellular Carcinoma.Asian Pac J Cancer Prev. 2023 Dec 1;24(12):4285-4292. doi: 10.31557/APJCP.2023.24.12.4285. Asian Pac J Cancer Prev. 2023. PMID: 38156865 Free PMC article.
-
Genome-wide interrogation of structural variation reveals novel African-specific prostate cancer oncogenic drivers.Genome Med. 2022 Aug 31;14(1):100. doi: 10.1186/s13073-022-01096-w. Genome Med. 2022. PMID: 36045381 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

