Single-cell mapping reveals new markers and functions of lymphatic endothelial cells in lymph nodes
- PMID: 32251437
- PMCID: PMC7162550
- DOI: 10.1371/journal.pbio.3000704
Single-cell mapping reveals new markers and functions of lymphatic endothelial cells in lymph nodes
Abstract
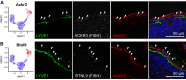
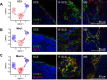
Lymph nodes (LNs) are highly organized secondary lymphoid organs that mediate adaptive immune responses to antigens delivered via afferent lymphatic vessels. Lymphatic endothelial cells (LECs) line intranodal lymphatic sinuses and organize lymph and antigen distribution. LECs also directly regulate T cells, mediating peripheral tolerance to self-antigens, and play a major role in many diseases, including cancer metastasis. However, little is known about the phenotypic and functional heterogeneity of LN LECs. Using single-cell RNA sequencing, we comprehensively defined the transcriptome of LECs in murine skin-draining LNs and identified new markers and functions of distinct LEC subpopulations. We found that LECs residing in the subcapsular sinus (SCS) have an unanticipated function in scavenging of modified low-density lipoprotein (LDL) and also identified a specific cortical LEC subtype implicated in rapid lymphocyte egress from LNs. Our data provide new, to our knowledge, insights into the diversity of LECs in murine LNs and a rich resource for future studies into the regulation of immune responses by LN LECs.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

