Recombinant protein production associated growth inhibition results mainly from transcription and not from translation
- PMID: 32252765
- PMCID: PMC7137236
- DOI: 10.1186/s12934-020-01343-y
Recombinant protein production associated growth inhibition results mainly from transcription and not from translation
Abstract
Background: Recombinant protein production can be stressful to the host organism. The extent of stress is determined by the specific properties of the recombinant transcript and protein, by the rates of transcription and translation, and by the environmental conditions encountered during the production process.
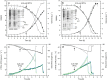
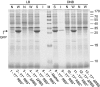
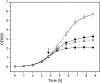
Results: The impact of the transcription of the T7-promoter controlled genes encoding human basic fibroblast growth factor (hFGF-2) and green fluorescent protein (GFP) as well as the translation into the recombinant protein on the growth properties of the production host E. coli BL21(DE3) were investigated. This was done by using expression vectors where the promoter region or the ribosome binding site(s) or both were removed. It is shown that already transcription without protein translation imposes a metabolic burden on the host cell. Translation of the transcript into large amounts of a properly folded protein does not show any effect on cell growth in the best case, e.g. high-level production of GFP in Luria-Bertani medium. However, translation appears to contribute to the metabolic burden if it is connected to protein folding associated problems, e.g. inclusion body formation.
Conclusion: The so-called metabolic burden of recombinant protein production is mainly attributed to transcription but can be enhanced through translation and those processes following translation (e.g. protein folding and degradation, heat-shock responses).
Keywords: Escherichia coli; Metabolic burden; Recombinant protein production; Transcriptional burden.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
-
- Bhattacharya SK, Dubey AK. Metabolic burden as reflected by maintenance coefficient of recombinant Escherichia coli overpressing target gene. Biotechnol Lett. 1995;17:1155–1160. doi: 10.1007/BF00128377. - DOI
-
- Lecina M, Sarro E, Casablancas A, Godia F, Cairo JJ. IPTG limitation avoids metabolic burden and acetic acid accumulation in induced fedbatch cultures of Escherichia coli M15 under glucose limiting conditions. Biochem Eng J. 2012;70:78–83. doi: 10.1016/j.bej.2012.10.006. - DOI
-
- Gasser B, Saloheimo M, Rinas U, Dragosits M, Rodríguez-Carmora E, Baumann K, Giuliani M, Parrilli E, Branduardi P, Lang C, Porro D, Ferrer P, Tutino ML, Mattanovich D, Villaverde A. Protein folding and conformational stress in microbial cells producing recombinant proteins: a host comparative overview. Microb Cell Fact. 2008;7:11. doi: 10.1186/1475-2859-7-11. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

