Characteristic features of the SERA multigene family in the malaria parasite
- PMID: 32252804
- PMCID: PMC7132891
- DOI: 10.1186/s13071-020-04044-y
Characteristic features of the SERA multigene family in the malaria parasite
Abstract
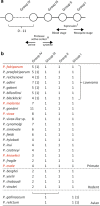
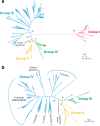
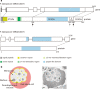
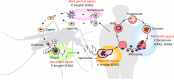
Serine repeat antigen (SERA) is conserved among species of the genus Plasmodium. Sera genes form a multigene family and are generally tandemly clustered on a single chromosome. Although all Plasmodium species encode multiple sera genes, the number varies between species. Among species, the members share similar sequences and gene organization. SERA possess a central papain-like cysteine protease domain, however, in some members, the active site cysteine residue is substituted with a serine. Recent studies implicate this gene family in a number of aspects in parasite biology and induction of protective immune response. This review summarizes the current understanding on this important gene family in several Plasmodium species. The Plasmodium falciparum (Pf)-sera family, for example, consists of nine gene members. Unlike other multigene families in Plasmodium species, Pf-sera genes do not exhibit antigenic variation. Pf-sera5 nucleotide diversity is also low. Moreover, although Pf-sera5 is highly transcribed during the blood stage of malaria infection, and a large amount is released into the host blood following schizont rupture, in malaria endemic countries the sero-positive rates for Pf-SERA5 are low, likely due to Pf-SERA5 binding of host proteins to avoid immune recognition. As an antigen, the N-terminal 47 kDa domain of Pf-SERA5 is a promising vaccine candidate currently undergoing clinical trials. Pf-SERA5 and Pf-SERA6, as well as P. berghei (Pb)-SERA3, and Pb-SERA5, have been investigated for their roles in parasite egress. Two P. yoelii SERA, which have a serine residue at the protease active center, are implicated in parasite virulence. Overall, these studies provide insight that during the evolution of the Plasmodium parasite, the sera gene family members have increased by gene duplication, and acquired various functions that enable the parasite to survive and successfully maintain infection in the host.
Keywords: Function; Gene family; Plasmodium; Polymorphism; SERA.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- WHO. World malaria report 2019. Geneva: World Health Organisation; 2019. https://www.who.int/news-room/feature-stories/detail/world-malaria-repor.... Accessed 19 Dec 2019.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

