Genomic Structure, Evolutionary Origins, and Reproductive Function of a Large Amplified Intrinsically Disordered Protein-Coding Gene on the X Chromosome (Laidx) in Mice
- PMID: 32253194
- PMCID: PMC7263670
- DOI: 10.1534/g3.120.401221
Genomic Structure, Evolutionary Origins, and Reproductive Function of a Large Amplified Intrinsically Disordered Protein-Coding Gene on the X Chromosome (Laidx) in Mice
Abstract
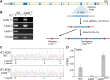
Mouse sex chromosomes are enriched for co-amplified gene families, present in tens to hundreds of copies. Co-amplification of Slx/Slxl1 on the X chromosome and Sly on the Y chromosome are involved in dose-dependent meiotic drive, however the role of other co-amplified genes remains poorly understood. Here we demonstrate that the co-amplified gene family on the X chromosome, Srsx, along with two additional partial gene annotations, is actually part of a larger transcription unit, which we name LaidxLaidx is harbored in a 229 kb amplicon that represents the ancestral state as compared to a 525 kb Y-amplicon containing the rearranged LaidyLaidx contains a 25,011 nucleotide open reading frame, predominantly expressed in round spermatids, predicted to encode an 871 kD protein. Laidx has orthologous copies with the rat and also the 825-MY diverged parasitic Chinese liver fluke, Clonorchis sinensis, the likely result of a horizontal gene transfer of rodent Laidx to an ancestor of the liver fluke. To assess the male reproductive functions of Laidx, we generated mice carrying a multi-megabase deletion of the Laidx-ampliconic region. Laidx-deficient male mice do not show detectable reproductive defects in fertility, fecundity, testis histology, and offspring sex ratio. We speculate that Laidx and Laidy represent a now inactive X vs. Y chromosome conflict that occurred in an ancestor of present day mice.
Keywords: Genetics of Sex; X chromosome; amplicon; horizontal gene transfer; male fertility; mouse; testicular germ cells.
Copyright © 2020 Arlt et al.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
