Mycoplasma bovis Membrane Protein MilA Is a Multifunctional Lipase with Novel Lipid and Glycosaminoglycan Binding Activity
- PMID: 32253247
- PMCID: PMC7240078
- DOI: 10.1128/IAI.00945-19
Mycoplasma bovis Membrane Protein MilA Is a Multifunctional Lipase with Novel Lipid and Glycosaminoglycan Binding Activity
Abstract
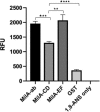
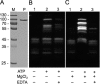
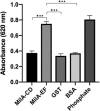
The survival, replication, and virulence of mycoplasmas depend on their ability to capture and import host-derived nutrients using poorly characterized membrane proteins. Previous studies on the important bovine pathogen Mycoplasma bovis demonstrated that the amino-terminal end of an immunogenic 226-kDa (P226) protein, encoded by milA (the full-length product of which has a predicted molecular weight of 303 kDa), had lipase activity. The predicted sequence of MilA contains glycosaminoglycan binding motifs, as well as multiple copies of a domain of unknown function (DUF445) that is also found in apolipoproteins. We mutagenized the gene to facilitate expression of a series of regions spanning the gene in Escherichia coli Using monospecific antibodies against these recombinant proteins, we showed that MilA was proteolytically processed into 226-kDa and 50-kDa fragments that were both partitioned into the detergent phase by Triton X-114 phase fractionation. Trypsin treatment of intact cells showed that P226 was surface exposed. In vitro, the recombinant regions of MilA bound to 1-anilinonaphthalene-8-sulfonic acid and to a variety of lipids. The MilA fragments were also shown to bind heparin. Antibody against the carboxyl-terminal fragment inhibited the growth of M. bovisin vitro This carboxyl end also bound and hydrolyzed ATP, suggestive of a potential role as an autotransporter. Our studies have demonstrated that DUF445 has lipid binding activity and that MilA is a multifunctional protein that may play multiple roles in the pathogenesis of infection with M. bovis.
Keywords: ATPase activity; GAG; Mycoplasma bovis; immunogenicity; lipid binding; membrane protein.
Copyright © 2020 American Society for Microbiology.
Figures





References
-
- Buchenau I, Poumarat F, Le Grand D, Linkner H, Rosengarten R, Hewicker-Trautwein M. 2010. Expression of Mycoplasma bovis variable surface membrane proteins in the respiratory tract of calves after experimental infection with a clonal variant of Mycoplasma bovis type strain PG45. Res Vet Sci 89:223–229. doi: 10.1016/j.rvsc.2010.03.014. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

