Dual Function of Aar, a Member of the New AraC Negative Regulator Family, in Escherichia coli Gene Expression
- PMID: 32253248
- PMCID: PMC7240098
- DOI: 10.1128/IAI.00100-20
Dual Function of Aar, a Member of the New AraC Negative Regulator Family, in Escherichia coli Gene Expression
Abstract
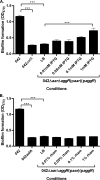
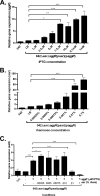
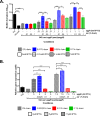
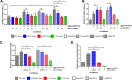
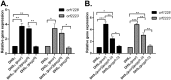
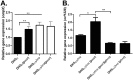
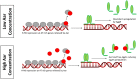
Enteroaggregative Escherichia coli (EAEC) is an E. coli pathotype associated with diarrhea and growth faltering. EAEC virulence gene expression is controlled by the autoactivated AraC family transcriptional regulator, AggR. AggR activates transcription of a large number of virulence genes, including Aar, which in turn acts as a negative regulator of AggR itself. Aar has also been shown to affect expression of E. coli housekeeping genes, including H-NS, a global regulator that acts at multiple promoters and silences AT-rich genes (such as those in the AggR regulon). Although Aar has been shown to bind both AggR and H-NS in vitro, functional significance of these interactions has not been shown in vivo In order to dissect this regulatory network, we removed the complex interdependence of aggR and aar by placing the genes under the control of titratable promoters. We measured phenotypic and genotypic changes on downstream genes in EAEC strain 042 and E. coli K-12 strain DH5α, which lacks the AggR regulon. In EAEC, we found that low expression of aar increases aafA fimbrial gene expression via H-NS; however, when aar is more highly expressed, it acts as a negative regulator via AggR. In DH5α, aar affected expression of E. coli genes in some cases via H-NS and in some cases independent of H-NS. Our data support the model that Aar interacts in concert with AggR, H-NS, and possibly other regulators and that these interactions are likely to be functionally significant in vivo.
Keywords: ANR; Aar; AggR; enteroaggregative E. coli.
Copyright © 2020 American Society for Microbiology.
Figures
References
-
- Mohamed JA, Huang DB, Jiang Z-D, DuPont HL, Nataro JP, Belkind-Gerson J, Okhuysen PC. 2007. Association of putative enteroaggregative Escherichia coli virulence genes and biofilm production in isolates from travelers to developing countries. J Clin Microbiol 45:121–126. doi: 10.1128/JCM.01128-06. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

