Integrating genomics for chickpea improvement: achievements and opportunities
- PMID: 32253478
- PMCID: PMC7214385
- DOI: 10.1007/s00122-020-03584-2
Integrating genomics for chickpea improvement: achievements and opportunities
Abstract
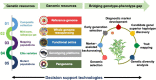
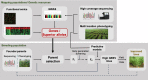
Integration of genomic technologies with breeding efforts have been used in recent years for chickpea improvement. Modern breeding along with low cost genotyping platforms have potential to further accelerate chickpea improvement efforts. The implementation of novel breeding technologies is expected to contribute substantial improvements in crop productivity. While conventional breeding methods have led to development of more than 200 improved chickpea varieties in the past, still there is ample scope to increase productivity. It is predicted that integration of modern genomic resources with conventional breeding efforts will help in the delivery of climate-resilient chickpea varieties in comparatively less time. Recent advances in genomics tools and technologies have facilitated the generation of large-scale sequencing and genotyping data sets in chickpea. Combined analysis of high-resolution phenotypic and genetic data is paving the way for identifying genes and biological pathways associated with breeding-related traits. Genomics technologies have been used to develop diagnostic markers for use in marker-assisted backcrossing programmes, which have yielded several molecular breeding products in chickpea. We anticipate that a sequence-based holistic breeding approach, including the integration of functional omics, parental selection, forward breeding and genome-wide selection, will bring a paradigm shift in development of superior chickpea varieties. There is a need to integrate the knowledge generated by modern genomics technologies with molecular breeding efforts to bridge the genome-to-phenome gap. Here, we review recent advances that have led to new possibilities for developing and screening breeding populations, and provide strategies for enhancing the selection efficiency and accelerating the rate of genetic gain in chickpea.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Similar articles
-
Genome-wide development and deployment of informative intron-spanning and intron-length polymorphism markers for genomics-assisted breeding applications in chickpea.Plant Sci. 2016 Nov;252:374-387. doi: 10.1016/j.plantsci.2016.08.013. Epub 2016 Aug 25. Plant Sci. 2016. PMID: 27717474
-
Chickpea (Cicer arietinum L.) battling against heat stress: plant breeding and genomics advances.Plant Mol Biol. 2025 Jul 31;115(4):101. doi: 10.1007/s11103-025-01628-z. Plant Mol Biol. 2025. PMID: 40745104 Review.
-
Translational genomics for achieving higher genetic gains in groundnut.Theor Appl Genet. 2020 May;133(5):1679-1702. doi: 10.1007/s00122-020-03592-2. Epub 2020 Apr 23. Theor Appl Genet. 2020. PMID: 32328677 Free PMC article. Review.
-
Genomics-assisted breeding for pigeonpea improvement.Theor Appl Genet. 2020 May;133(5):1721-1737. doi: 10.1007/s00122-020-03563-7. Epub 2020 Feb 15. Theor Appl Genet. 2020. PMID: 32062675 Review.
-
Current advances in chickpea genomics: applications and future perspectives.Plant Cell Rep. 2018 Jul;37(7):947-965. doi: 10.1007/s00299-018-2305-6. Epub 2018 Jun 2. Plant Cell Rep. 2018. PMID: 29860584 Review.
Cited by
-
Genetic mapping of QTLs for drought tolerance in chickpea (Cicer arietinum L.).Front Genet. 2022 Aug 19;13:953898. doi: 10.3389/fgene.2022.953898. eCollection 2022. Front Genet. 2022. PMID: 36061197 Free PMC article.
-
Genetic variation in CaTIFY4b contributes to drought adaptation in chickpea.Plant Biotechnol J. 2022 Sep;20(9):1701-1715. doi: 10.1111/pbi.13840. Epub 2022 May 21. Plant Biotechnol J. 2022. PMID: 35534989 Free PMC article.
-
Editorial: Accelerating Genetic Gains in Pulses.Front Plant Sci. 2022 Apr 7;13:879377. doi: 10.3389/fpls.2022.879377. eCollection 2022. Front Plant Sci. 2022. PMID: 35463449 Free PMC article. No abstract available.
-
Advances in "Omics" Approaches for Improving Toxic Metals/Metalloids Tolerance in Plants.Front Plant Sci. 2022 Jan 4;12:794373. doi: 10.3389/fpls.2021.794373. eCollection 2021. Front Plant Sci. 2022. PMID: 35058954 Free PMC article. Review.
-
High confidence QTLs and key genes identified using Meta-QTL analysis for enhancing heat tolerance in chickpea (Cicer arietinum L.).Front Plant Sci. 2023 Oct 20;14:1274759. doi: 10.3389/fpls.2023.1274759. eCollection 2023. Front Plant Sci. 2023. PMID: 37929162 Free PMC article.
References
-
- Abbo S, Bonfil DJ, Berkovitch Z, Reifen R. Towards enhancing lutein concentration in chickpea, cultivar and management effects. Plant Breed. 2010;129:407–411.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

