Using Direct RNA Nanopore Sequencing to Deconvolute Viral Transcriptomes
- PMID: 32255550
- PMCID: PMC7187905
- DOI: 10.1002/cpmc.99
Using Direct RNA Nanopore Sequencing to Deconvolute Viral Transcriptomes
Abstract
The genomes of DNA viruses encode deceptively complex transcriptomes evolved to maximize coding potential within the confines of a relatively small genome. Defining the full range of viral RNAs produced during an infection is key to understanding the viral replication cycle and its interactions with the host cell. Traditional short-read (Illumina) sequencing approaches are problematic in this setting due to the difficulty of assigning short reads to individual RNAs in regions of transcript overlap and to the biases introduced by the required recoding and amplification steps. Additionally, different methodologies may be required to analyze the 5' and 3' ends of RNAs, which increases both cost and effort. The advent of long-read nanopore sequencing simplifies this approach by providing a single assay that captures and sequences full length RNAs, either in cDNA or native RNA form. The latter is particularly appealing as it reduces known recoding biases whilst allowing more advanced analyses such as estimation of poly(A) tail length and the detection of RNA modifications including N6 -methyladenosine. Using herpes simplex virus (HSV)-infected primary fibroblasts as a template, we provide a step-by-step guide to the production of direct RNA sequencing libraries suitable for sequencing using Oxford Nanopore Technologies platforms and provide a simple computational approach to deriving a high-quality annotation of the HSV transcriptome from the resulting sequencing data. © 2020 by John Wiley & Sons, Inc. Basic Protocol 1: Productive infection of primary fibroblasts with herpes simplex virus Support Protocol: Cell passage and plating of primary fibroblasts Basic Protocol 2: Preparation and sequencing of dRNA-seq libraries from virus-infected cells Basic Protocol 3: Processing, alignment, and analysis of dRNA-seq datasets.
Keywords: DNA virus; RNA sequencing; annotation; herpesvirus; nanopore; viral transcriptome.
© 2020 John Wiley & Sons, Inc.
Conflict of interest statement
Conflict of Interest
The authors declare no conflicts of interest.
Figures

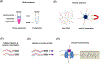
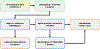
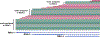
References
Literature cited
-
- Arias C, Weisburd B, Stern-Ginossar N, Mercier A, Madrid AS, Bellare P, … Ganem D (2014). KSHV 2.0: A Comprehensive Annotation of the Kaposi’s Sarcoma-Associated Herpesvirus Genome Using Next-Generation Sequencing Reveals Novel Genomic and Functional Features. PLoS Pathogens, 10(1). 10.1371/journal.ppat.1003847 - DOI - PMC - PubMed
-
- Chomczynski P (1993). A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. BioTechniques, 15(3), 532–537. - PubMed
Key References
-
- Garalde DR, Snell EA, Jachimowicz D, Sipos B, Lloyd JH, Bruce M, Pantic N, Admassu T, James P, Warland A, Jordan M, Ciccone J, Serra S, Keenan J, Martin S, McNeill L, Wallace EJ, Jayasinghe L, Wright C, … Turner DJ (2018). Highly parallel direct RNA sequencing on an array of nanopores. Nature Methods, 15(3), 201–206. 10.1038/nmeth.4577 - DOI - PubMed
-
- First application of dRNA-seq using nanopore arrays. Provides useful discussion of the strengths and limitations of the technique and highlights the potential for detection of modified bases.
-
- A short review discussing the utility of the major RNA-seq methodologies to the analysis of viral transcriptomes.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

