Ion Mobility and Gas-Phase Covalent Labeling Study of the Structure and Reactivity of Gaseous Ubiquitin Ions Electrosprayed from Aqueous and Denaturing Solutions
- PMID: 32255627
- PMCID: PMC7205579
- DOI: 10.1021/jasms.9b00138
Ion Mobility and Gas-Phase Covalent Labeling Study of the Structure and Reactivity of Gaseous Ubiquitin Ions Electrosprayed from Aqueous and Denaturing Solutions
Abstract
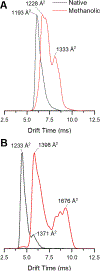
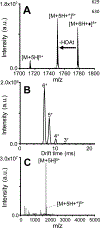
Gas-phase ion/ion chemistry was coupled to ion mobility/mass spectrometry analysis to correlate the structure of gaseous ubiquitin to its solution structures with selective covalent structural probes. Collision cross section (CCS) distributions were measured to ensure the ubiquitin ions were not unfolded when they were introduced to the gas phase. Aqueous solutions stabilizing the native state of ubiquitin yielded folded ubiquitin structures with CCS values consistent with previously published literature. Denaturing solutions favored several families of unfolded conformations for most of the charge states evaluated. Gas-phase covalent labeling via ion/ion reactions was followed by collision-induced dissociation of the intact, labeled protein to determine which residues were labeled. Ubiquitin 5+ and 6+ electrosprayed from aqueous conditions were covalently modified preferentially at the lysine 29 and arginine 54 positions, indicating that elements of three-dimensional structure were maintained in the gas phase. On the other hand, most ubiquitin ions produced in denaturing conditions were labeled at various other lysine residues, likely due to the availability of additional sites following methanol- and low-pH-induced unfolding. These data support the conservation of ubiquitin structural elements in the gas phase. The research presented here provides the basis for residue-specific characterization of biomolecules in the gas phase.
Keywords: covalent labeling; ion mobility; ion/ion reactions; native mass spectrometry.
Figures

References
-
- Ishima R; Torchia DA, Protein dynamics from NMR. Nat Struct Biol 2000, 7 (9), 740–3. - PubMed
-
- Loo JA; Loo RRO; Udseth HR; Edmonds CG; Smith RD, Solvent-Induced Conformational-Changes of Polypeptides Probed by Electrospray-Ionization Mass-Spectrometry. Rapid Commun Mass Sp 1991, 5 (3), 101–105. - PubMed
-
- Chowdhury SK; Katta V; Chait BT, An electrospray-ionization mass spectrometer with new features. Rapid Commun Mass Spectrom 1990, 4 (3), 81–7. - PubMed
-
- Ishii K; Zhou M; Uchiyama S, Native mass spectrometry for understanding dynamic protein complex. Biochim Biophys Acta Gen Subj 2018, 1862 (2), 275–286. - PubMed
-
- Lanucara F; Holman SW; Gray CJ; Eyers CE, The power of ion mobility-mass spectrometry for structural characterization and the study of conformational dynamics. Nat Chem 2014, 6 (4), 281–294. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

