Functional Genomics of the Pediatric Obese Asthma Phenotype Reveal Enrichment of Rho-GTPase Pathways
- PMID: 32255672
- PMCID: PMC7365356
- DOI: 10.1164/rccm.201906-1199OC
Functional Genomics of the Pediatric Obese Asthma Phenotype Reveal Enrichment of Rho-GTPase Pathways
Abstract
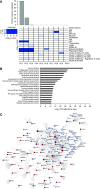
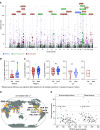
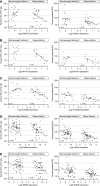
Rationale: Obesity-related asthma disproportionately affects minority children and is associated with nonatopic T-helper type 1 (Th1) cell polarized inflammation that correlates with pulmonary function deficits. Its underlying mechanisms are poorly understood.Objectives: To use functional genomics to identify cellular mechanisms associated with nonatopic inflammation in obese minority children with asthma.Methods: CD4+ (cluster of differentiation 4-positive) Th cells from 59 obese Hispanic and African American children with asthma and 61 normal-weight Hispanic and African American children with asthma underwent quantification of the transcriptome and DNA methylome and genotyping. Expression and methylation quantitative trait loci revealed the contribution of genetic variation to transcription and DNA methylation. Adjusting for Th-cell subtype proportions discriminated loci where transcription or methylation differences were driven by differences in subtype proportions from loci that were independently associated with obesity-related asthma.Measurements and Main Results: Obese children with asthma had more memory and fewer naive Th cells than normal-weight children with asthma. Differentially expressed and methylated genes and methylation quantitative trait loci in obese children with asthma, independent of Th-cell subtype proportions, were enriched in Rho-GTPase pathways. Inhibition of CDC42 (cell division cycle 42), one of the Rho-GTPases associated with Th-cell differentiation, was associated with downregulation of the IFNγ, but not the IL-4, gene. Differential expression of the RPS27L (40S ribosomal protein S27-like) gene, part of the p53/mammalian target of rapamycin pathway, was due to nonrandom distribution of expression quantitative trait loci variants between groups. Differentially expressed and/or methylated genes, including RPS27L, were associated with pulmonary function deficits in obese children with asthma.Conclusions: We found enrichment of Rho-GTPase pathways in obese asthmatic Th cells, identifying them as a novel therapeutic target for obesity-related asthma, a disease that is suboptimally responsive to current therapies.
Keywords: DNA methylation; children; expression and methylation quantitative trait loci; gene expression; obesity-related asthma.
Figures






Comment in
-
A Role for the Rho-GTPase Pathway in Pediatric Obese Asthma.Am J Respir Crit Care Med. 2020 Jul 15;202(2):169-170. doi: 10.1164/rccm.202004-1073ED. Am J Respir Crit Care Med. 2020. PMID: 32479737 Free PMC article. No abstract available.
References
-
- Rastogi D, Canfield SM, Andrade A, Isasi CR, Hall CB, Rubinstein A, et al. Obesity-associated asthma in children: a distinct entity. Chest. 2012;141:895–905. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

