In situ estimation of genetic variation of functional and ecological traits in Quercus petraea and Q.robur
- PMID: 32256274
- PMCID: PMC7136077
- DOI: 10.1007/s11295-019-1407-9
In situ estimation of genetic variation of functional and ecological traits in Quercus petraea and Q.robur
Abstract
Background: Predicting the evolutionary potential of natural tree populations requires the estimation of heritability and genetic correlations among traits on which selection acts, as differences in evolutionary success between species may rely on differences for these genetic parameters. In situ estimates are expected to be more accurate than measures done under controlled conditions which do not reflect the natural environmental variance.
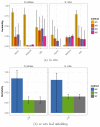
Aims: The aim of the current study was to estimate three genetic parameters (i.e. heritability, evolvability and genetic correlations) in a natural mixed oak stand composed of Quercus petraea and Quercus robur about 100 years old, for 58 traits of ecological and functional relevance (growth, reproduction, phenology, physiology, resilience, structure, morphology and defence).
Methods: First we estimated genetic parameters directly in situ using realized genomic relatedness of adult trees and parentage relationships over two generations to estimate the traits additive variance. Secondly, we benefited from existing ex situ experiments (progeny tests and conservation collection) installed with the same populations, thus allowing comparisons of in situ heritability estimates with more traditional methods.
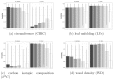
Results: Heritability and evolvability estimates obtained with different methods varied substantially and showed large confidence intervals, however we found that in situ were less precise than ex situ estimates, and assessments over two generations (with deeper relatedness) improved estimates of heritability while large sampling sizes are needed for accurate estimations. At the biological level, heritability values varied moderately across different ecological and functional categories of traits, and genetic correlations among traits were conserved over the two species.
Conclusion: We identified limits for using realized genomic relatedness in natural stands to estimate the genetic variance, given the overall low variance of genetic relatedness and the rather low sampling sizes of currently used long term genetic plots in forestry. These limits can be overcome if larger sample sizes are considered, or if the approach is extended over the next generation.
Keywords: Heritability; evolvability; genetic correlation; genomic relatedness; natural population; tree.
Conflict of interest statement
Conflict of interest disclosure The authors of this preprint declare that they have no financial conflict of interest with the content of this article.
Figures





References
-
- Bacilieri R, Ducousso A, Kremer A. Comparison of morphological characters and molecular markers for the analysis of hybridization in sessile and pedunculate oak. Annales des Sciences Forestieres. 1996;53:79–91.
-
- Bacilieri R, Labbe T, Kremer A. Intraspecific genetic structure in a mixed population of Quercus petraea (Matt.) Leibl and Q. roburL. Heredity. 1994;73:130–141.
Grants and funding
LinkOut - more resources
Full Text Sources
