Crucial Role of the Accessory Genome in the Evolutionary Trajectory of Acinetobacter baumannii Global Clone 1
- PMID: 32256462
- PMCID: PMC7093585
- DOI: 10.3389/fmicb.2020.00342
Crucial Role of the Accessory Genome in the Evolutionary Trajectory of Acinetobacter baumannii Global Clone 1
Abstract
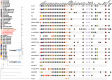
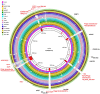
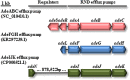
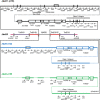
Acinetobacter baumannii is one of the most important nosocomial pathogens able to rapidly develop extensive drug resistance. Here, we study the role of accessory genome in the success of the globally disseminated clone 1 (GC1) with functional and genomic approaches. Comparative genomics was performed with available GC1 genomes (n = 106) against other A. baumannii high-risk and sporadic clones. Genetic traits related to accessory genome were found common and conserved along time as two novel regions of genome plasticity, and a CRISPR-Cas system acquired before clonal diversification located at the same loci as "sedentary" modules. Although identified within hotspot for recombination, other block of accessory genome was also "sedentary" in lineage 1 of GC1 with signs of microevolution as the AbaR0-type genomic island (GI) identified in A144 and in A155 strains which were maintained one month in independent experiments without antimicrobial pressure. The prophage YMC/09/02/B1251_ABA_BP was found to be "mobile" since, although it was shared by all GC1 genomes, it showed high intrinsic microevolution as well as mobility to different insertion sites. Interestingly, a wide variety of Insertion Sequences (IS), probably acquired by the flow of plasmids related to Rep_3 superfamily was found. These IS showed dissimilar genomic location amongst GC1 genomes presumably associated with promptly niche adaptation. On the other hand, a type VI secretion system and three efflux pumps were subjected to deep processes of genomic loss in A. baumannii but not in GC1. As a whole, these findings suggest that preservation of some genetic modules of accessory genome harbored by strains from different continents in combination with great plasticity of IS and varied flow of plasmids, may be central features of the genomic structure of GC1. Competition of A144 and A155 versus A118 (ST 404/ND) without antimicrobial pressure suggested a higher ability of GC1 to grow over a clone with sporadic behavior which explains, from an ecological perspective, the global achievement of this successful pandemic clone in the hospital habitat. Together, these data suggest an essential role of still unknown properties of "mobile" and "sedentary" accessory genome that is preserved over time under different antibiotic or stress conditions.
Keywords: A. baumannii; accessory genome; genomic plasticity; high-risk clone; international clone 1 (GC1).
Copyright © 2020 Álvarez, Quiroga, Galán, Vilacoba, Quiroga, Ramírez and Centrón.
Figures

References
-
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. - PubMed
-
- Antunes L. C. S., Imperi F., Towner K. J., Visca P. (2011). Genome-assisted identification of putative iron-utilization genes in Acinetobacter baumannii and their distribution among a genotypically diverse collection of clinical isolates. Res. Microbiol. 162 279–284. 10.1016/j.resmic.2010.10.010 - DOI - PubMed
-
- Arduino S. M., Quiroga M. P., Ramírez M. S., Merkier A. K., Errecalde L., Di Martino A., et al. (2012). Transposons and integrons in colistin-resistant clones of Klebsiella pneumoniae and Acinetobacter baumannii with epidemic or sporadic behaviour. J. Med. Microbiol. 61 1417–1420. 10.1099/jmm.0.038968-0 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

