Haplotype-Based Analysis of KIR-Gene Profiles in a South European Population-Distribution of Standard and Variant Haplotypes, and Identification of Novel Recombinant Structures
- PMID: 32256494
- PMCID: PMC7089957
- DOI: 10.3389/fimmu.2020.00440
Haplotype-Based Analysis of KIR-Gene Profiles in a South European Population-Distribution of Standard and Variant Haplotypes, and Identification of Novel Recombinant Structures
Abstract
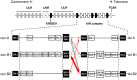
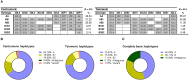
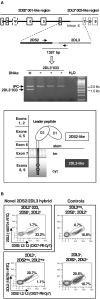
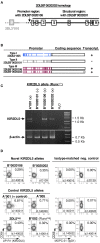
Inhibitory Killer-cell Immunoglobulin-like Receptors (KIR) specific for HLA class I molecules enable human natural killer cells to monitor altered antigen presentation in pathogen-infected and tumor cells. KIR genes display extensive copy-number variation and allelic polymorphism. They organize in a series of variable arrangements, designated KIR haplotypes, which derive from duplications of ancestral genes and sequence diversification through point mutation and unequal crossing-over events. Genomic studies have established the organization of multiple KIR haplotypes-many of them are fixed in most human populations, whereas variants of those have less certain distributions. Whilst KIR-gene diversity of many populations and ethnicities has been explored superficially (frequencies of individual genes and presence/absence profiles), less abundant are in-depth analyses of how such diversity emerges from KIR-haplotype structures. We characterize here the genetic diversity of KIR in a sample of 414 Spanish individuals. Using a parsimonious approach, we manage to explain all 38 observed KIR-gene profiles by homo- or heterozygous combinations of six fixed centromeric and telomeric motifs; of six variant gene arrangements characterized previously by us and others; and of two novel haplotypes never detected before in Caucasoids. Associated to the latter haplotypes, we also identified the novel transcribed KIR2DL5B*0020202 allele, and a chimeric KIR2DS2/KIR2DL3 gene (designated KIR2DL3*033) that challenges current criteria for classification and nomenclature of KIR genes and haplotypes.
Keywords: KIR; NK cells; copy-number variation; genes; haplotypes; polymorphism.
Copyright © 2020 Cisneros, Moraru, Gómez-Lozano, Muntasell, López-Botet and Vilches.
Figures



Similar articles
-
Allele-level haplotype frequencies and pairwise linkage disequilibrium for 14 KIR loci in 506 European-American individuals.PLoS One. 2012;7(11):e47491. doi: 10.1371/journal.pone.0047491. Epub 2012 Nov 5. PLoS One. 2012. PMID: 23139747 Free PMC article.
-
Killer-cell immunoglobulin-like receptor (KIR) and human leukocyte antigen (HLA) class I genetic diversity in four South African populations.Hum Immunol. 2017 Jul-Aug;78(7-8):503-509. doi: 10.1016/j.humimm.2017.05.006. Epub 2017 May 29. Hum Immunol. 2017. PMID: 28571758
-
Recombinant structures expand and contract inter and intragenic diversification at the KIR locus.BMC Genomics. 2013 Feb 8;14:89. doi: 10.1186/1471-2164-14-89. BMC Genomics. 2013. PMID: 23394822 Free PMC article.
-
The extensive polymorphism of KIR genes.Immunology. 2010 Jan;129(1):8-19. doi: 10.1111/j.1365-2567.2009.03208.x. Immunology. 2010. PMID: 20028428 Free PMC article. Review.
-
Variation within the human killer cell immunoglobulin-like receptor (KIR) gene family.Crit Rev Immunol. 2002;22(5-6):463-82. Crit Rev Immunol. 2002. PMID: 12803322 Review.
Cited by
-
A Protective HLA Extended Haplotype Outweighs the Major COVID-19 Risk Factor Inherited From Neanderthals in the Sardinian Population.Front Immunol. 2022 Apr 19;13:891147. doi: 10.3389/fimmu.2022.891147. eCollection 2022. Front Immunol. 2022. PMID: 35514995 Free PMC article.
-
Donor selection for adoptive immunotherapy with NK cells in AML patients: Comparison between analysis of lytic NK cell clones and phenotypical identification of alloreactive NK cell repertoire.Front Immunol. 2023 Feb 14;14:1111419. doi: 10.3389/fimmu.2023.1111419. eCollection 2023. Front Immunol. 2023. PMID: 36865545 Free PMC article.
-
Epitope characterization of a monoclonal antibody that selectively recognizes KIR2DL1 allotypes.HLA. 2022 Aug;100(2):107-118. doi: 10.1111/tan.14630. Epub 2022 Jun 9. HLA. 2022. PMID: 35411634 Free PMC article.
-
Immunogenomics of Killer Cell Immunoglobulin-Like Receptor (KIR) and HLA Class I: Coevolution and Consequences for Human Health.J Allergy Clin Immunol Pract. 2022 Jul;10(7):1763-1775. doi: 10.1016/j.jaip.2022.04.036. Epub 2022 May 10. J Allergy Clin Immunol Pract. 2022. PMID: 35561968 Free PMC article.
-
KIR Allelic Variation and the Remission of Atopic Dermatitis Over Time.Immunohorizons. 2023 Jan 1;7(1):30-40. doi: 10.4049/immunohorizons.2200095. Immunohorizons. 2023. PMID: 36637513 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

