Complete Chloroplast Genome Sequence of Hibiscus cannabinus and Comparative Analysis of the Malvaceae Family
- PMID: 32256523
- PMCID: PMC7090147
- DOI: 10.3389/fgene.2020.00227
Complete Chloroplast Genome Sequence of Hibiscus cannabinus and Comparative Analysis of the Malvaceae Family
Abstract
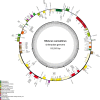
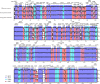
Kenaf (Hibiscus cannabinus) is one of the most fast-growing bast in the world and belongs to the family Malvaceae. However, the systematic classification and chloroplast (cp) genome of kenaf has not been reported to date. In this study, we sequenced the cp genome of kenaf and conducted phylogenetic and comparative analyses in the family of Malvaceae. The sizes of H. cannabinus cp genomes were 162,903 bp in length, containing 113 unique genes (79 protein-coding genes, four rRNA genes, and 30 tRNA genes). Phylogenetic analysis indicated that the cp genome sequence of H. cannabinus has closer relationships with Talipariti hamabo and Abelmoschus esculentus than with Hibiscus syriacus, which disagrees with the taxonomical relationship. Further analysis obtained a new version of the cp genome annotation of H. syriacus and found that the orientation variation of small single copy (SSC) region exists widely in the family of Malvaceae. The highly variable ycf1 and the highly conserved gene rrn32 were identified among the family of Malvaceae. In particular, the explanation for two different SSC orientations in the cp genomes associated with phylogenetic analysis is discussed. These results provide insights into the systematic classification of the Hibiscus genus in the Malvaceae family.
Keywords: Hibiscus cannabinus; LSC; Malvaceae; SSC; cp; gene orientation.
Copyright © 2020 Cheng, Zhang, Qi and Zhang.
Figures



Similar articles
-
Complete chloroplast genome of a semi-mangrove plant Hibiscus tiliaceus (Malvaceae).Mitochondrial DNA B Resour. 2021 Jun 14;6(7):1904-1905. doi: 10.1080/23802359.2021.1935337. Mitochondrial DNA B Resour. 2021. PMID: 34179470 Free PMC article.
-
The complete chloroplast genome of Hibiscus Taiwanensis (Malvaceae).Mitochondrial DNA B Resour. 2019 Jul 13;4(2):2533-2534. doi: 10.1080/23802359.2019.1640084. Mitochondrial DNA B Resour. 2019. PMID: 33365613 Free PMC article.
-
Comparative analysis of chloroplast genomes of kenaf cytoplasmic male sterile line and its maintainer line.Sci Rep. 2021 Mar 5;11(1):5301. doi: 10.1038/s41598-021-84567-1. Sci Rep. 2021. PMID: 33674697 Free PMC article.
-
The complete chloroplast genome of Malvaviscus penduliflorus (Malvaceae).Mitochondrial DNA B Resour. 2023 Aug 21;8(8):886-889. doi: 10.1080/23802359.2023.2246670. eCollection 2023. Mitochondrial DNA B Resour. 2023. PMID: 37614528 Free PMC article. Review.
-
Kenaf (Hibiscus cannabinus L.) Seed and its Potential Food Applications: A Review.J Food Sci. 2019 Aug;84(8):2015-2023. doi: 10.1111/1750-3841.14714. Epub 2019 Jul 30. J Food Sci. 2019. PMID: 31364175 Review.
Cited by
-
Black queen cell virus detected in Canadian mosquitoes.J Insect Sci. 2023 Mar 1;23(2):10. doi: 10.1093/jisesa/iead016. J Insect Sci. 2023. PMID: 37004145 Free PMC article.
-
Revisiting Phryma leptostachya L.: phylogenetic relationships and biogeographical patterns from complete plastome.BMC Plant Biol. 2025 Mar 4;25(1):278. doi: 10.1186/s12870-025-06272-9. BMC Plant Biol. 2025. PMID: 40033209 Free PMC article.
-
Multiple domestication events explain the origin of Gossypium hirsutum landraces in Mexico.Ecol Evol. 2023 Mar 8;13(3):e9838. doi: 10.1002/ece3.9838. eCollection 2023 Mar. Ecol Evol. 2023. PMID: 36911302 Free PMC article.
-
Unfolding the Complete Chloroplast Genome of Myrica esculenta Buch.-Ham. ex D.Don (1825): Advancing Phylogenetic Insights Within Fagales and Pioneering DNA Barcodes for Precise Species Identification.Ecol Evol. 2025 Jun 12;15(6):e71566. doi: 10.1002/ece3.71566. eCollection 2025 Jun. Ecol Evol. 2025. PMID: 40519893 Free PMC article.
-
Disjunction and Vicariance Between East and West Asia: A Case Study on Euonymus sect. Uniloculares Based on Plastid Genome Analysis.Front Plant Sci. 2022 Mar 11;13:825209. doi: 10.3389/fpls.2022.825209. eCollection 2022. Front Plant Sci. 2022. PMID: 35360330 Free PMC article.
References
-
- Cheng Z., Lu B.-R., Sameshima K., Fu D.-X., Chen J.-K. (2004). Identification and genetic relationships of kenaf (Hibiscus cannabinus L.) germplasm revealed by AFLP analysis. Genet. Resourc. Crop Evol. 51 393–401. 10.1023/b:gres.0000023454.96401.1c - DOI
-
- Christenhusz M. J. M., Byng J. W. (2016). The number of known plants species in the world and its annual increase. Phytotaxa 261:201 10.11646/phytotaxa.261.3.1 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

