Somatic Mutations in HER2 and Implications for Current Treatment Paradigms in HER2-Positive Breast Cancer
- PMID: 32256585
- PMCID: PMC7081042
- DOI: 10.1155/2020/6375956
Somatic Mutations in HER2 and Implications for Current Treatment Paradigms in HER2-Positive Breast Cancer
Abstract
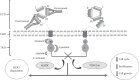
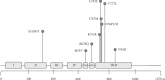
In one of every four or five cases of breast cancer, the human epidermal growth factor receptor-2 (HER2) gene is overexpressed. These carcinomas are known as HER2-positive. HER2 overexpression is linked to an aggressive phenotype and a lower rate of disease-free and overall survival. Drugs such as trastuzumab, pertuzumab, lapatinib, neratinib, and the more recent afatinib target the deregulation of HER2 expression. Some authors have attributed somatic mutations in HER2, a role in resistance to anti-HER2 therapy as differential regulation of HER2 has been observed among patients. Recently, studies in metastatic ER + tumors suggest that some HER2 mutations emerge as a mechanism of acquired resistance to endocrine therapy. In an effort to identify possible biomarkers of the efficacy of anti-HER2 therapy, we here review the known single-nucleotide polymorphisms (SNPs) of the HER2 gene found in HER2-positive breast cancer patients and their relationship with clinical outcomes. Information was recompiled on 11 somatic HER2 SNPs. Seven polymorphisms are located in the tyrosine kinase domain region of the gene contrasting with the low number of mutations found in extracellular and transmembrane areas. HER2-positive patients carrying S310F, S310Y, R678Q, D769H, or I767M mutations seem good candidates for anti-HER2 therapy as they show favorable outcomes and a good response to current pharmacological treatments. Carrying the L755S or D769Y mutation could also confer benefits when receiving neratinib or afatinib. By contrast, patients with mutations L755S, V842I, K753I, or D769Y do not seem to benefit from trastuzumab. Resistance to lapatinib has been reported in patients with L755S, V842I, and K753I. These data suggest that exploring HER2 SNPs in each patient could help individualize anti-HER2 therapies. Advances in our understanding of the genetics of the HER2 gene and its relations with the efficacy of anti-HER2 treatments are needed to improve the outcomes of patients with this aggressive breast cancer.
Copyright © 2020 Maria Gaibar et al.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

