Cell proliferation is induced in renal cell carcinoma through miR-92a-3p upregulation by targeting FBXW7
- PMID: 32256821
- PMCID: PMC7074420
- DOI: 10.3892/ol.2020.11443
Cell proliferation is induced in renal cell carcinoma through miR-92a-3p upregulation by targeting FBXW7
Abstract
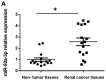
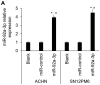
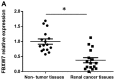
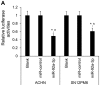
Renal cell carcinoma (RCC) is the most common type of kidney cancer whose incidence has gradually increased worldwide. MicroRNAs (miRNAs) represent a type of short endogenous non-coding RNA containing approximately 22 nucleotides, which are capable of regulating mRNAs at the post-transcriptional level in human cells. miRNAs have been demonstrated to mediate gene expression by influencing important regulatory genes. Accumulating evidence indicates that certain miRNAs are involved in RCC development. The present study investigated the underlying mechanism and functional role of miR-92a-3p in RCC cells using reverse transcription-quantitative polymerase chain reaction, western blotting, 3' UTR luciferase assay, cell proliferation assay and soft agar assay. The results demonstrated that miR-92a-3p expression level is significantly upregulated in RCC tissues and cell lines; however, F-box and WD repeat domain containing 7 (FBXW7) expression level was significantly downregulated in RCC tissues and cell lines. Subsequently, whether FBXW7 could be considered as a direct target of miR-92a-3p in RCC cells was investigated. The results demonstrated that miR-92a-3p overexpression significantly promoted RCC cell proliferation and colony formation. Conversely, miR-92a-3p downregulation significantly inhibited RCC cell proliferation and colony formation. In addition, FBXW7 knockdown significantly enhanced RCC cell proliferation and colony formation. Conversely, FBXW7 overexpression significantly inhibited RCC cell proliferation and colony formation. Collectively, these results demonstrated that miR-92a-3p/FBXW7 pathway may represent a novel strategy and therapeutic target for RCC.
Keywords: F-box and WD repeat domain containing 7; anchorage-independent growth; cell proliferation; microRNA-92a-3p.
Copyright: © Zeng et al.
Figures

























References
-
- Zhou J, Yun EJ, Chen W, Ding Y, Wu K, Wang B, Ding C, Hernandez E, Santoyo J, Pong RC, et al. Targeting 3-phosphoinositide-dependent protein kinase 1 associated with drug-resistant renal cell carcinoma using new oridonin analogs. Cell Death Dis. 2017;8:e2701. doi: 10.1038/cddis.2017.121. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
