hPCL3S promotes proliferation and migration of androgen-independent prostate cancer cells
- PMID: 32256978
- PMCID: PMC7105160
- DOI: 10.18632/oncotarget.27511
hPCL3S promotes proliferation and migration of androgen-independent prostate cancer cells
Abstract
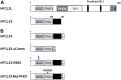
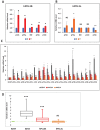
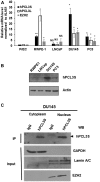
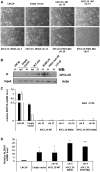
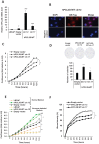
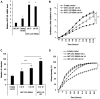
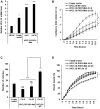
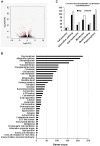
Polycomb repressive complex 2 (PRC2) allows the deposition of H3K27me3. PRC2 facultative subunits modulate its activity and recruitment such as hPCL3/PHF19, a human ortholog of Drosophila Polycomb-like protein (PCL). These proteins contain a TUDOR domain binding H3K36me3, two PHD domains and a "Winged-helix" domain involved in GC-rich DNA binding. The human PCL3 locus encodes the full-length hPCL3L protein and a shorter isoform, hPCL3S containing the TUDOR and PHD1 domains only. In this study, we demonstrated by RT-qPCR analyses of 25 prostate tumors that hPCL3S is frequently up-regulated. In addition, hPCL3S is overexpressed in the androgen-independent DU145 and PC3 cells, but not in the androgen-dependent LNCaP cells. hPCL3S knockdown decreased the proliferation and migration of DU145 and PC3 whereas its forced expression into LNCaP increased these properties. A mutant hPCL3S unable to bind H3K36me3 (TUDOR-W50A) increased proliferation and migration of LNCaP similarly to wt hPCL3S whereas inactivation of its PHD1 domain decreased proliferation. These effects partially relied on the up-regulation of genes known to be important for the proliferation and/or migration of prostate cancer cells such as S100A16, PlexinA2, and Spondin1. Collectively, our results suggest hPCL3S as a new potential therapeutic target in castration resistant prostate cancers.
Keywords: PHF19; PRC2; hPCL3S; prostate cancer; β-catenin.
Conflict of interest statement
CONFLICTS OF INTEREST The authors declare no disclosure of potential conflicts of interest.
Figures




References
LinkOut - more resources
Full Text Sources
Miscellaneous

