PreMSIm: An R package for predicting microsatellite instability from the expression profiling of a gene panel in cancer
- PMID: 32257050
- PMCID: PMC7113609
- DOI: 10.1016/j.csbj.2020.03.007
PreMSIm: An R package for predicting microsatellite instability from the expression profiling of a gene panel in cancer
Abstract
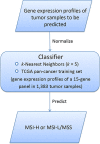
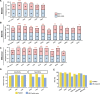
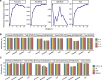
Microsatellite instability (MSI) is a genomic property of the cancers with defective DNA mismatch repair and is a useful marker for cancer diagnosis and treatment in diverse cancer types. In particular, MSI has been associated with the active immune checkpoint blockade therapy response in cancer. Most of computational methods for predicting MSI are based on DNA sequencing data and a few are based on mRNA expression data. Using the RNA-Seq pan-cancer datasets for three cancer cohorts (colon, gastric, and endometrial cancers) from The Cancer Genome Atlas (TCGA) program, we developed an algorithm (PreMSIm) for predicting MSI from the expression profiling of a 15-gene panel in cancer. We demonstrated that PreMSIm had high prediction performance in predicting MSI in most cases using both RNA-Seq and microarray gene expression datasets. Moreover, PreMSIm displayed superior or comparable performance versus other DNA or mRNA-based methods. We conclude that PreMSIm has the potential to provide an alternative approach for identifying MSI in cancer.
Keywords: ACC, adrenocortical carcinoma; AUC, area under the curve; Algorithm; BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; CV, cross validation; Cancer; Classification; DLBC, lymphoid neoplasm diffuse large B-cell lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; GEO, Gene Expression Omnibus; GO, gene ontology; Gene expression profiling; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LAML, acute myeloid leukemia; LGG, brain lower grade glioma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MESO, mesothelioma; MSI, microsatellite instability; MSS, microsatellite stability; Machine learning; Microsatellite instability; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PPI, protein-protein interaction; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; RF, random forest; ROC, receiver operating characteristic; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; SVM, support vector machine; TCGA, The Cancer Genome Atlas; TGCT, testicular germ cell tumors; THCA, thyroid carcinoma; THYM, thymoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterine carcinosarcoma; UVM, uveal melanoma; XGBoost, extreme gradient boosting; k-NN, k-nearest neighbor.
© 2020 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


Similar articles
-
Identification of SHCBP1 as a potential biomarker involving diagnosis, prognosis, and tumor immune microenvironment across multiple cancers.Comput Struct Biotechnol J. 2022 Jun 18;20:3106-3119. doi: 10.1016/j.csbj.2022.06.039. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35782736 Free PMC article.
-
Violations of proportional hazard assumption in Cox regression model of transcriptomic data in TCGA pan-cancer cohorts.Comput Struct Biotechnol J. 2022 Jan 7;20:496-507. doi: 10.1016/j.csbj.2022.01.004. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35070171 Free PMC article.
-
APOBEC3C is a novel target for the immune treatment of lower-grade gliomas.Neurol Res. 2024 Mar;46(3):227-242. doi: 10.1080/01616412.2023.2287340. Epub 2024 Jan 22. Neurol Res. 2024. PMID: 38007705
-
The cancer driver genes IDH1/2, JARID1C/ KDM5C, and UTX/ KDM6A: crosstalk between histone demethylation and hypoxic reprogramming in cancer metabolism.Exp Mol Med. 2019 Jun 20;51(6):1-17. doi: 10.1038/s12276-019-0230-6. Exp Mol Med. 2019. PMID: 31221981 Free PMC article. Review.
-
Good or not good: Role of miR-18a in cancer biology.Rep Pract Oncol Radiother. 2020 Sep-Oct;25(5):808-819. doi: 10.1016/j.rpor.2020.07.006. Epub 2020 Aug 12. Rep Pract Oncol Radiother. 2020. PMID: 32884453 Free PMC article. Review.
Cited by
-
Zic Family Member 2 (ZIC2): a Potential Diagnostic and Prognostic Biomarker for Pan-Cancer.Front Mol Biosci. 2021 Feb 16;8:631067. doi: 10.3389/fmolb.2021.631067. eCollection 2021. Front Mol Biosci. 2021. PMID: 33665207 Free PMC article.
-
Integrated Analysis of Copy Number Variation, Microsatellite Instability, and Tumor Mutation Burden Identifies an 11-Gene Signature Predicting Survival in Breast Cancer.Front Cell Dev Biol. 2021 Sep 28;9:721505. doi: 10.3389/fcell.2021.721505. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34650974 Free PMC article.
-
Cancer-specific functional profiling in microsatellite-unstable (MSI) colon and endometrial cancers using combined differentially expressed genes and biclustering analysis.Medicine (Baltimore). 2023 May 12;102(19):e33647. doi: 10.1097/MD.0000000000033647. Medicine (Baltimore). 2023. PMID: 37171359 Free PMC article.
-
The Molecular Effects of a High Fat Diet on Endometrial Tumour Biology.Life (Basel). 2020 Sep 10;10(9):188. doi: 10.3390/life10090188. Life (Basel). 2020. PMID: 32927694 Free PMC article.
-
DNA damage response in breast cancer and its significant role in guiding novel precise therapies.Biomark Res. 2024 Sep 27;12(1):111. doi: 10.1186/s40364-024-00653-2. Biomark Res. 2024. PMID: 39334297 Free PMC article. Review.
References
-
- Hegde M. ACMG technical standards and guidelines for genetic testing for inherited colorectal cancer (Lynch syndrome, familial adenomatous polyposis, and MYH-associated polyposis) Genet Med. 2014;16(1):101–116. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
