Molecular mapping of dominant gene responsible for leaf curl virus resistance in chilli pepper (Capsicum annuum L.)
- PMID: 32257738
- PMCID: PMC7103024
- DOI: 10.1007/s13205-020-02168-7
Molecular mapping of dominant gene responsible for leaf curl virus resistance in chilli pepper (Capsicum annuum L.)
Erratum in
-
Correction to: Molecular mapping of dominant gene responsible for leaf curl virus resistance in chilli pepper (Capsicum annuum L.).3 Biotech. 2020 Oct;10(10):417. doi: 10.1007/s13205-020-02402-2. Epub 2020 Sep 4. 3 Biotech. 2020. PMID: 32953379 Free PMC article.
Abstract
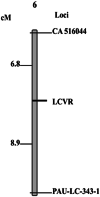
A resistant source (S-343) having monogenic dominant resistance to chilli leaf curl virus disease (ChiLCVD) has been identified at Punjab Agricultural University (PAU), Ludhiana. The F2 mapping population of 204 plants was derived from the cross MS-341 (susceptible) × S-343 (resistant) to identify the linked marker with the disease-resistant gene. Out of the 685 single-sequence repeats (SSRs) used, only 160 primers showed parental polymorphism. These 160 polymorphic primers were used for bulk segregant analysis and only eight SSR primers were able to differentiate the resistant and susceptible bulks. The linkage analysis revealed that the two markers CA 516044 and PAU-LC-343-1 were found linked with the disease-resistant gene covering a total distance of 15.7 centimorgan (cM). The two primers CA 516044 and PAU-LC-343-1 were found located on chromosome 6 of the pepper genome at a genetic distance of 6.8 cM and 8.9 cM, respectively, from the resistant gene. The validation of linked markers was performed using 26 resistant and susceptible genotypes developed at PAU, Ludhiana by former researchers. The validation of the primers revealed that there was a correlation between phenotypic and genotypic data of the used genotypes, and these markers can be used for the marker-assisted breeding procedures for transferring ChiLCVD resistance until the gene-based markers will be developed. The markers described in this study are the first-ever molecular markers identified as linked to the ChiLCVD-resistant gene.
Keywords: Capsicum annuum; Dominant gene; Leaf curl virus; Molecular mapping; Resistance; SSRs.
© King Abdulaziz City for Science and Technology 2020.
Conflict of interest statement
Conflict of interestThe authors declare no competing interests.
Figures







References
-
- Ahmad A, Sharma A, Zehra SB, Kang SS, Bhat M, Hussain A. Evaluation of chilli genotypes against Chilli leaf curl virus. Indian J Ecol. 2016;43:144–147.
-
- Aulakh PS, Dhaliwal MS, Jindal SK, Schafleitner R, Singh K. Mapping of male sterility gene ms10 in chilli pepper (Capsicum annuum L.) Plant Breed. 2016;135:531–535. doi: 10.1111/pbr.12389. - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

