Methodological Insight Into Mosquito Microbiome Studies
- PMID: 32257962
- PMCID: PMC7089923
- DOI: 10.3389/fcimb.2020.00086
Methodological Insight Into Mosquito Microbiome Studies
Abstract
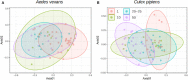
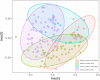
Symbiotic bacteria affect competence for pathogen transmission in insect vectors, including mosquitoes. However, knowledge on mosquito-microbiome-pathogen interactions remains limited, largely due to methodological reasons. The current, cost-effective practice of sample pooling used in mosquito surveillance and epidemiology prevents correlation of individual traits (i.e., microbiome profile) and infection status. Moreover, many mosquito studies employ laboratory-reared colonies that do not necessarily reflect the natural microbiome composition and variation in wild populations. As a consequence, epidemiological and microbiome studies in mosquitoes are to some extent uncoupled, and the interactions among pathogens, microbiomes, and natural mosquito populations remain poorly understood. This study focuses on the effect the pooling practice poses on mosquito microbiome profiles, and tests different approaches to find an optimized low-cost methodology for extensive sampling while allowing for accurate, individual-level microbiome studies. We tested the effect of pooling by comparing wild-caught, individually processed mosquitoes with pooled samples. With individual mosquitoes, we also tested two methodological aspects that directly affect the cost and feasibility of broad-scale molecular studies: sample preservation and tissue dissection. Pooling affected both alpha- and beta-diversity measures of the microbiome, highlighting the importance of using individual samples when possible. Both RNA and DNA yields were higher when using inexpensive reagents such as NAP (nucleic acid preservation) buffer or absolute ethanol, without freezing for short-term storage. Microbiome alpha- and beta-diversity did not show overall significant differences between the tested treatments compared to the controls (freshly extracted samples or dissected guts). However, the use of standardized protocols is highly recommended to avoid methodological bias in the data.
Keywords: dissection; epidemiology; microbiome; mosquito; pooling; preservation; vector.
Copyright © 2020 Rodríguez-Ruano, Juhaňáková, Vávra and Nováková.
Figures



References
-
- Astrid T., Margit E., Leopold F. (2016). Ethanol: a simple and effective RNA-preservation for freshwater insects living in remote habitats. Limnol. Oceanogr. Methods 14, 186–195. 10.1002/lom3.10079 - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

