Distinct Roles of TRAPPC8 and TRAPPC12 in Ciliogenesis via Their Interactions With OFD1
- PMID: 32258032
- PMCID: PMC7090148
- DOI: 10.3389/fcell.2020.00148
Distinct Roles of TRAPPC8 and TRAPPC12 in Ciliogenesis via Their Interactions With OFD1
Abstract
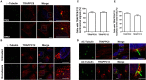
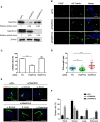
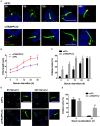
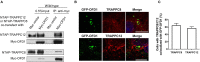
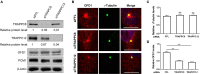
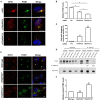
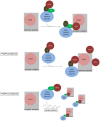
The transport protein particle (TRAPP) complex was initially identified as a tethering factor for COPII vesicle. Subsequently, three forms (TRAPPI, II, and III) have been found and TRAPPIII has been reported to serve as a regulator in autophagy. This study investigates a new role of mammalian TRAPPIII in ciliogenesis. We found a ciliopathy protein, oral-facial-digital syndrome 1 (OFD1), interacting with the TRAPPIII-specific subunits TRAPPC8 and TRAPPC12. TRAPPC8 is necessary for the association of OFD1 with pericentriolar material 1 (PCM1). Its depletion reduces the extent of colocalized signals between OFD1 and PCM1, but does not compromise the structural integrity of centriolar satellites. The interaction between TRAPPC8 and OFD1 inhibits that between OFD1 and TRAPPC12, suggesting different roles of TRAPPIII-specific subunits in ciliogenesis and explaining the differences in cilium lengths in TRAPPC8-depleted and TRAPPC12-depleted hTERT-RPE1 cells. On the other hand, TRAPPC12 depletion causes increased ciliary length because TRAPPC12 is required for the disassembly of primary cilia. Overall, this study has revealed different roles of TRAPPC8 and TRAPPC12 in the assembly of centriolar satellites and demonstrated a possible tethering role of TRAPPIII during ciliogenesis.
Keywords: OFD1; TRAPPC12; TRAPPC8; centriolar satellite; primary cilium.
Copyright © 2020 Zhang, Li, Siu, Luo and Yu.
Figures


Similar articles
-
Mammalian TRAPPIII Complex positively modulates the recruitment of Sec13/31 onto COPII vesicles.Sci Rep. 2017 Feb 27;7:43207. doi: 10.1038/srep43207. Sci Rep. 2017. PMID: 28240221 Free PMC article.
-
Autophagy promotes primary ciliogenesis by removing OFD1 from centriolar satellites.Nature. 2013 Oct 10;502(7470):254-7. doi: 10.1038/nature12606. Epub 2013 Oct 2. Nature. 2013. PMID: 24089205 Free PMC article.
-
The adaptor function of TRAPPC2 in mammalian TRAPPs explains TRAPPC2-associated SEDT and TRAPPC9-associated congenital intellectual disability.PLoS One. 2011;6(8):e23350. doi: 10.1371/journal.pone.0023350. Epub 2011 Aug 15. PLoS One. 2011. PMID: 21858081 Free PMC article.
-
Autophagy and Ciliogenesis.JMA J. 2021 Jul 15;4(3):207-215. doi: 10.31662/jmaj.2021-0090. Epub 2021 Jul 6. JMA J. 2021. PMID: 34414314 Free PMC article. Review.
-
Current topics of functional links between primary cilia and cell cycle.Cilia. 2015 Dec 29;4:12. doi: 10.1186/s13630-015-0021-1. eCollection 2015. Cilia. 2015. PMID: 26719793 Free PMC article. Review.
Cited by
-
The TRAPP complexes: oligomeric exchange factors that activate the small GTPases Rab1 and Rab11.FEBS Lett. 2023 Mar;597(6):734-749. doi: 10.1002/1873-3468.14553. Epub 2022 Dec 18. FEBS Lett. 2023. PMID: 36477798 Free PMC article. Review.
-
Myosin VI regulates ciliogenesis by promoting the turnover of the centrosomal/satellite protein OFD1.EMBO Rep. 2022 Feb 3;23(3):e54160. doi: 10.15252/embr.202154160. Epub 2021 Dec 27. EMBO Rep. 2022. PMID: 34957672 Free PMC article.
-
TRAPPopathies: Severe Multisystem Disorders Caused by Variants in Genes of the Transport Protein Particle (TRAPP) Complexes.Int J Mol Sci. 2024 Dec 12;25(24):13329. doi: 10.3390/ijms252413329. Int J Mol Sci. 2024. PMID: 39769094 Free PMC article. Review.
-
Circadian oscillation in primary cilium length by clock genes regulates fibroblast cell migration.EMBO Rep. 2023 Dec 6;24(12):e56870. doi: 10.15252/embr.202356870. Epub 2023 Nov 16. EMBO Rep. 2023. PMID: 37971148 Free PMC article.
-
TRAPPC2l Participates in Male Germ Cell Development by Regulating Cell Division.Cell Prolif. 2025 Jun;58(6):e13810. doi: 10.1111/cpr.13810. Epub 2025 Jan 26. Cell Prolif. 2025. PMID: 40539230 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials

