Evolution and Developmental System Drift in the Endoderm Gene Regulatory Network of Caenorhabditis and Other Nematodes
- PMID: 32258041
- PMCID: PMC7093329
- DOI: 10.3389/fcell.2020.00170
Evolution and Developmental System Drift in the Endoderm Gene Regulatory Network of Caenorhabditis and Other Nematodes
Abstract
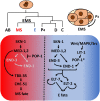
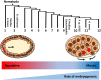
Developmental gene regulatory networks (GRNs) underpin metazoan embryogenesis and have undergone substantial modification to generate the tremendous variety of animal forms present on Earth today. The nematode Caenorhabditis elegans has been a central model for advancing many important discoveries in fundamental mechanistic biology and, more recently, has provided a strong base from which to explore the evolutionary diversification of GRN architecture and developmental processes in other species. In this short review, we will focus on evolutionary diversification of the GRN for the most ancient of the embryonic germ layers, the endoderm. Early embryogenesis diverges considerably across the phylum Nematoda. Notably, while some species deploy regulative development, more derived species, such as C. elegans, exhibit largely mosaic modes of embryogenesis. Despite the relatively similar morphology of the nematode gut across species, widespread variation has been observed in the signaling inputs that initiate the endoderm GRN, an exemplar of developmental system drift (DSD). We will explore how genetic variation in the endoderm GRN helps to drive DSD at both inter- and intraspecies levels, thereby resulting in a robust developmental system. Comparative studies using divergent nematodes promise to unveil the genetic mechanisms controlling developmental plasticity and provide a paradigm for the principles governing evolutionary modification of an embryonic GRN.
Keywords: Caenorhabditis; developmental hourglass; developmental system drift; plasticity; robustness.
Copyright © 2020 Ewe, Torres Cleuren and Rothman.
Figures
References
-
- Alcorn M. R., Callander D. C., López-Santos A., Cleuren Y. N. T., Birsoy B., Joshi P. M., et al. (2016). Heterotaxy in Caenorhabditis: widespread natural variation in left – right arrangement of the major organs. Philos. Trans. R. Soc. B Biol. Sci. 371:20150404. 10.1098/rstb.2015.0404 - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

