Gene Expression Analysis of Human Papillomavirus-Associated Colorectal Carcinoma
- PMID: 32258125
- PMCID: PMC7103040
- DOI: 10.1155/2020/5201587
Gene Expression Analysis of Human Papillomavirus-Associated Colorectal Carcinoma
Abstract
Purpose: Human papillomavirus (HPV) antigens had been found in colorectal cancer (CRC) tissue, but little evidence demonstrates the association of HPV with oncogene mutations in CRC. We aim to elucidate the mutated genes that link HPV infection and CRC carcinogenesis.
Methods: Cancerous and adjacent noncancerous tissues were obtained from CRC patients. HPV antigen was measured by using the immunohistochemical (IHC) technique. The differentially expressed genes (DEGs) in HPV-positive and HPV-negative tumor tissues were measured by using TaqMan Array Plates. The target genes were validated with the qPCR method.
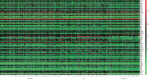
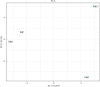
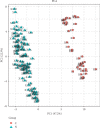
Results: 15 (31.9%) cases of CRC patients were observed to be HPV positive, in which HPV antigen was expressed in most tumor tissues rather than in adjacent noncancerous tissues. With TaqMan Array Plates analyses, we found that 39 differentially expressed genes (DEGs) were upregulated, while 17 DEGs were downregulated in HPV-positive CRC tissues compared with HPV-negative tissues. Four DEGs (MMP-7, MYC, WNT-5A, and AXIN2) were upregulated in tumor vs. normal tissues, or adenoma vs. normal tissue in TCGA, which was overlapped with our data. In the confirmation test, MMP-7, MYC, WNT-5A, and AXIN2 were upregulated in cancerous tissue compared with adjacent noncancerous tissue. MYC, WNT-5A, and AXIN2 were shown to be upregulated in HPV-positive CRC tissues when compared to HPV-negative tissues.
Conclusion: HPV-encoding genome may integrate into the tumor genomes that involved in multiple signaling pathways. Further genomic and proteomic investigation is necessary for obtaining a more comprehensive knowledge of signaling pathways associated with the CRC carcinogenesis.
Copyright © 2020 Qiancheng Qiu et al.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Detection of HPV and the role of p16INK4A overexpression as a surrogate marker for the presence of functional HPV oncoprotein E7 in colorectal cancer.BMC Cancer. 2010 Mar 26;10:117. doi: 10.1186/1471-2407-10-117. BMC Cancer. 2010. PMID: 20346145 Free PMC article.
-
Detection of Human papillomavirus and the role of p16INK4a in colorectal carcinomas.PLoS One. 2020 Jun 25;15(6):e0235065. doi: 10.1371/journal.pone.0235065. eCollection 2020. PLoS One. 2020. PMID: 32584870 Free PMC article.
-
Human papillomavirus infection correlates with inflammatory Stat3 signaling activity and IL-17 level in patients with colorectal cancer.PLoS One. 2015 Feb 23;10(2):e0118391. doi: 10.1371/journal.pone.0118391. eCollection 2015. PLoS One. 2015. PMID: 25706309 Free PMC article.
-
Prevalence of human papillomavirus in Chinese patients with colorectal cancer.Colorectal Dis. 2011 Aug;13(8):865-71. doi: 10.1111/j.1463-1318.2010.02335.x. Epub 2010 May 28. Colorectal Dis. 2011. PMID: 20528894
-
Human papillomavirus and colorectal cancer: evidences and pitfalls of published literature.Int J Colorectal Dis. 2011 Feb;26(2):135-42. doi: 10.1007/s00384-010-1049-8. Epub 2010 Sep 1. Int J Colorectal Dis. 2011. PMID: 20809427 Review.
Cited by
-
Microbiome Differences in Colorectal Cancer Patients and Healthy Individuals: Implications for Vaccine Antigen Discovery.Immunotargets Ther. 2024 Dec 13;13:749-774. doi: 10.2147/ITT.S486731. eCollection 2024. Immunotargets Ther. 2024. PMID: 39698218 Free PMC article. Review.
-
WNT5a in Colorectal Cancer: Research Progress and Challenges.Cancer Manag Res. 2021 Mar 16;13:2483-2498. doi: 10.2147/CMAR.S289819. eCollection 2021. Cancer Manag Res. 2021. PMID: 33758546 Free PMC article. Review.
-
Colorectal cancer and gut viruses: a visualized analysis based on CiteSpace knowledge graph.Front Microbiol. 2023 Oct 18;14:1239818. doi: 10.3389/fmicb.2023.1239818. eCollection 2023. Front Microbiol. 2023. PMID: 37928670 Free PMC article.
-
Association of miRNA targetome variants in LAMC1 and GNB3 genes with colorectal cancer and obesity.Cancer Med. 2022 Nov;11(21):3923-3938. doi: 10.1002/cam4.4713. Epub 2022 Apr 4. Cancer Med. 2022. PMID: 35373932 Free PMC article.
-
Gut microbiota: Role and Association with Tumorigenesis in Different Malignancies.Mol Biol Rep. 2022 Aug;49(8):8087-8107. doi: 10.1007/s11033-022-07357-6. Epub 2022 May 11. Mol Biol Rep. 2022. PMID: 35543828 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

