Homology Modeling of Human Uridine-5'-diphosphate-glucuronosyltransferase 1A6 Reveals Insights into Factors Influencing Substrate and Cosubstrate Binding
- PMID: 32258923
- PMCID: PMC7114752
- DOI: 10.1021/acsomega.0c00205
Homology Modeling of Human Uridine-5'-diphosphate-glucuronosyltransferase 1A6 Reveals Insights into Factors Influencing Substrate and Cosubstrate Binding
Abstract
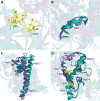
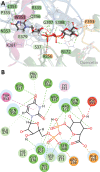
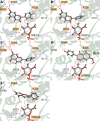
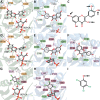
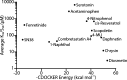
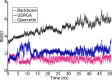
The elimination of numerous endogenous compounds and xenobiotics via glucuronidation by uridine-5'-diphosphate glycosyltransferase enzymes (UGTs) is an essential process of the body's chemical defense system. UGTs have distinct but overlapping substrate preferences, but the molecular basis for their substrate specificity remains poorly understood. Three-dimensional protein structures can greatly enhance our understanding of the interactions between enzymes and their substrates, but because of the inherent difficulties in purifying and crystallizing integral endoplasmic reticulum membrane proteins, no complete mammalian UGT structure has yet been produced. To address this problem, we have created a homology model of UGT1A6 using I-TASSER to explore, in detail, the interactions of human UGT1A6 with its substrates. Ligands were docked into our model in the presence of the cosubstrate uridine-5'-diphosphate-glucuronic acid, interacting residues were examined, and poses were compared to those cocrystallized with various plant and bacterial glycosyltransferases (GTs). Our model structurally resembles other GTs, and docking experiments replicated many of the expected UGT-substrate interactions. Some bias toward the template structures' protein-substrate interactions and binding preferences was evident.
Copyright © 2020 American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

Similar articles
-
Insights into the UDP-sugar selectivities of human UDP-glycosyltransferases (UGT): a molecular modeling perspective.Drug Metab Rev. 2015 Aug;47(3):335-45. doi: 10.3109/03602532.2015.1071835. Epub 2015 Aug 3. Drug Metab Rev. 2015. PMID: 26289097 Review.
-
Inhibitory effects of uridine diphosphate on UDP-glucuronosyltransferase.Life Sci. 1998;63(19):1693-9. doi: 10.1016/s0024-3205(98)00441-x. Life Sci. 1998. PMID: 9806225
-
In silico insights: chemical and structural characteristics associated with uridine diphosphate-glucuronosyltransferase substrate selectivity.Clin Exp Pharmacol Physiol. 2003 Nov;30(11):836-40. doi: 10.1046/j.1440-1681.2003.03923.x. Clin Exp Pharmacol Physiol. 2003. PMID: 14678246
-
Structure and Protein-Protein Interactions of Human UDP-Glucuronosyltransferases.Front Pharmacol. 2016 Oct 24;7:388. doi: 10.3389/fphar.2016.00388. eCollection 2016. Front Pharmacol. 2016. PMID: 27822186 Free PMC article. Review.
-
[Crystal structures of plant uridine diphosphate-dependent glycosyltransferases].Sheng Wu Gong Cheng Xue Bao. 2014 Jun;30(6):838-47. Sheng Wu Gong Cheng Xue Bao. 2014. PMID: 25212002 Review. Chinese.
Cited by
-
Subfunctionalization of a monolignol to a phytoalexin glucosyltransferase is accompanied by substrate inhibition.Plant Commun. 2023 May 8;4(3):100506. doi: 10.1016/j.xplc.2022.100506. Epub 2022 Dec 24. Plant Commun. 2023. PMID: 36566353 Free PMC article.
-
Curcumin in Cancer Prevention: Insights from Clinical Trials and Strategies to Enhance Bioavailability.Curr Pharm Des. 2024;30(23):1838-1851. doi: 10.2174/0113816128303514240517054617. Curr Pharm Des. 2024. PMID: 38808709 Review.
-
A potential implication of UDP-glucuronosyltransferase 2B10 in the detoxification of drugs used in pediatric hematopoietic stem cell transplantation setting: an in silico investigation.BMC Mol Cell Biol. 2022 Jan 21;23(1):5. doi: 10.1186/s12860-021-00402-5. BMC Mol Cell Biol. 2022. PMID: 35062878 Free PMC article.
-
Similarities in Structure and Function of UDP-Glycosyltransferase Homologs from Human and Plants.Int J Mol Sci. 2024 Feb 28;25(5):2782. doi: 10.3390/ijms25052782. Int J Mol Sci. 2024. PMID: 38474028 Free PMC article. Review.
-
Structural and functional studies reveal the molecular basis of substrate promiscuity of a glycosyltransferase originating from a major agricultural pest.J Biol Chem. 2023 Dec;299(12):105421. doi: 10.1016/j.jbc.2023.105421. Epub 2023 Nov 1. J Biol Chem. 2023. PMID: 37923139 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

