Genome-Wide RNAi Screen Identifies Regulators of Cardiomyocyte Necrosis
- PMID: 32259070
- PMCID: PMC7088983
- DOI: 10.1021/acsptsci.9b00052
Genome-Wide RNAi Screen Identifies Regulators of Cardiomyocyte Necrosis
Abstract
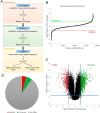
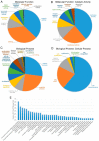
Regulation of cellular death is central to nearly all physiological routines and is dysregulated in virtually all diseases. Cell death occurs by two major processes, necrosis which culminates in a pervasive inflammatory response and apoptosis which is largely immunologically inert. As necrosis has long been considered an accidental, unregulated form of cellular death that occurred in response to a harsh environmental stimulus, it was largely ignored as a clinical target. However, recent elegant studies suggest that certain forms of necrosis can be reprogrammed. However, scant little is known about the molecules and pathways that orchestrate calcium-overload-induced necrosis, a main mediator of ischemia/reperfusion (IR)-induced cardiomyocyte cell death. To rectify this critical gap in our knowledge, we performed a novel genome-wide siRNA screen to identify modulators of calcium-induced necrosis in human muscle cells. Our screen identified multiple molecular circuitries that either enhance or inhibit this process, including lysosomal calcium channel TPCN1, mitophagy mediatorTOMM7, Ran-binding protein RanBP9, Histone deacetylase HDAC2, chemokine CCL11, and the Arp2/3 complex regulator glia maturation factor-γ (GMFG). Notably, a number of druggable enzymes were identified, including the proteasome β5 subunit (encoded by PSMB5 gene), which controls the proteasomal chymotrypsin-like peptidase activity. Such findings open up the possibility for the discovery of pharmacological interventions that could provide therapeutic benefits to patients affected by myriad disorders characterized by excessive (or too little) necrotic cell loss, including but not limited to IR injury in the heart and kidney, chronic neurodegenerative disorders, muscular dystrophies, sepsis, and cancers.
Copyright © 2019 American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
