Coexpression of FOXP3 and a Helios isoform enhances the effectiveness of human engineered regulatory T cells
- PMID: 32259202
- PMCID: PMC7160257
- DOI: 10.1182/bloodadvances.2019000965
Coexpression of FOXP3 and a Helios isoform enhances the effectiveness of human engineered regulatory T cells
Abstract
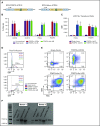
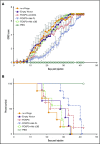
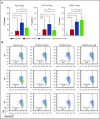
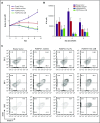
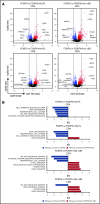
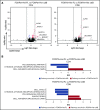
Regulatory T cells (Tregs) are a subset of immune cells that suppress the immune response. Treg therapy for inflammatory diseases is being tested in the clinic, with moderate success. However, it is difficult to isolate and expand Tregs to sufficient numbers. Engineered Tregs (eTregs) can be generated in larger quantities by genetically manipulating conventional T cells to express FOXP3. These eTregs can suppress in vitro and in vivo but not as effectively as endogenous Tregs. We hypothesized that ectopic expression of the transcription factor Helios along with FOXP3 is required for optimal eTreg immunosuppression. To test this theory, we generated eTregs by retrovirally transducing total human T cells (CD4+ and CD8+) with FOXP3 alone or with each of the 2 predominant isoforms of Helios. Expression of both FOXP3 and the full-length isoform of Helios was required for eTreg-mediated disease delay in a xenogeneic graft-versus-host disease model. In vitro, this corresponded with superior suppressive function of FOXP3 and full-length Helios-expressing CD4+ and CD8+ eTregs. RNA sequencing showed that the addition of full-length Helios changed gene expression in cellular pathways and the Treg signature compared with FOXP3 alone or the other major Helios isoform. Together, these results show that functional human CD4+ and CD8+ eTregs can be generated from total human T cells by coexpressing FOXP3 and full-length Helios.
© 2020 by The American Society of Hematology.
Conflict of interest statement
Conflict-of-interest disclosure: M.A.M. is a consultant for Johnson & Johnson Global Services for topics unrelated to this work. M.A.M., A.S., R.T.F., and T.M.Y. have filed a patent provisional based on the work in this publication. The remaining authors declare no competing financial interests.
Figures


References
-
- Sakaguchi S, Sakaguchi N, Asano M, Itoh M, Toda M. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J Immunol. 1995;155(3):1151-1164. - PubMed
-
- Brunkow ME, Jeffery EW, Hjerrild KA, et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet. 2001;27(1):68-73. - PubMed
-
- Bennett CL, Christie J, Ramsdell F, et al. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet. 2001;27(1):20-21. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

