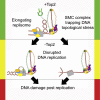
Cohesin Causes Replicative DNA Damage by Trapping DNA Topological Stress
- PMID: 32259483
- PMCID: PMC7242899
- DOI: 10.1016/j.molcel.2020.03.013
Cohesin Causes Replicative DNA Damage by Trapping DNA Topological Stress
Abstract
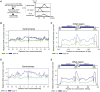
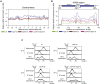
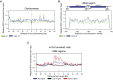
DNA topological stress inhibits DNA replication fork (RF) progression and contributes to DNA replication stress. In Saccharomyces cerevisiae, we demonstrate that centromeric DNA and the rDNA array are especially vulnerable to DNA topological stress during replication. The activity of the SMC complexes cohesin and condensin are linked to both the generation and repair of DNA topological-stress-linked damage in these regions. At cohesin-enriched centromeres, cohesin activity causes the accumulation of DNA damage, RF rotation, and pre-catenation, confirming that cohesin-dependent DNA topological stress impacts on normal replication progression. In contrast, at the rDNA, cohesin and condensin activity inhibit the repair of damage caused by DNA topological stress. We propose that, as well as generally acting to ensure faithful genetic inheritance, SMCs can disrupt genome stability by trapping DNA topological stress.
Keywords: DNA damage; DNA replication stress; DNA topology; SMC; cohesin; condensin; topoisomerases.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures


References
-
- Aguilera A., García-Muse T. Causes of genome instability. Annu. Rev. Genet. 2013;47:1–32. - PubMed
-
- Akai Y., Kurokawa Y., Nakazawa N., Tonami-Murakami Y., Suzuki Y., Yoshimura S.H., Iwasaki H., Shiroiwa Y., Nakamura T., Shibata E., Yanagida M. Opposing role of condensin hinge against replication protein A in mitosis and interphase through promoting DNA annealing. Open Biol. 2011;1:110023. - PMC - PubMed
-
- Aragón L. The Smc5/6 complex: new and old functions of the enigmatic long-distance relative. Annu. Rev. Genet. 2018;52:89–107. - PubMed
-
- Baxter J., Diffley J.F. Topoisomerase II inactivation prevents the completion of DNA replication in budding yeast. Mol. Cell. 2008;30:790–802. - PubMed
-
- Baxter J., Sen N., Martínez V.L., De Carandini M.E.M., Schvartzman J.B., Diffley J.F., Aragón L. Positive supercoiling of mitotic DNA drives decatenation by topoisomerase II in eukaryotes. Science. 2011;331:1328–1332. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

