Genetic Screen for Cell Fitness in High or Low Oxygen Highlights Mitochondrial and Lipid Metabolism
- PMID: 32259488
- PMCID: PMC7293541
- DOI: 10.1016/j.cell.2020.03.029
Genetic Screen for Cell Fitness in High or Low Oxygen Highlights Mitochondrial and Lipid Metabolism
Abstract
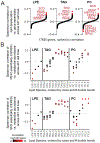
Human cells are able to sense and adapt to variations in oxygen levels. Historically, much research in this field has focused on hypoxia-inducible factor (HIF) signaling and reactive oxygen species (ROS). Here, we perform genome-wide CRISPR growth screens at 21%, 5%, and 1% oxygen to systematically identify gene knockouts with relative fitness defects in high oxygen (213 genes) or low oxygen (109 genes), most without known connection to HIF or ROS. Knockouts of many mitochondrial pathways thought to be essential, including complex I and enzymes in Fe-S biosynthesis, grow relatively well at low oxygen and thus are buffered by hypoxia. In contrast, in certain cell types, knockout of lipid biosynthetic and peroxisomal genes causes fitness defects only in low oxygen. Our resource nominates genetic diseases whose severity may be modulated by oxygen and links hundreds of genes to oxygen homeostasis.
Keywords: CoQ biosynthesis; FASII; MPC; TMEM189; hypoxia; iron-sulfur clusters; membrane fluidity; plasmalogens; pyruvate dehydrogenase; type II fatty acid synthesis.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests V.K.M. is a paid scientific advisor to 5AM Ventures and Janssen Pharmaceuticals. O.S.S. is a paid consultant for Proteinaceous. V.K.M. and I.H.J are listed as inventors on a patent application filed by Massachusetts General on the use of hypoxia as a therapeutic strategy.
Figures

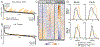



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

