Nuclear hubs built on RNAs and clustered organization of the genome
- PMID: 32259767
- PMCID: PMC7371543
- DOI: 10.1016/j.ceb.2020.02.015
Nuclear hubs built on RNAs and clustered organization of the genome
Abstract
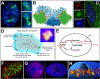
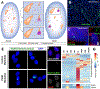
RNAs play diverse roles in formation and function of subnuclear compartments, most of which are associated with active genes. NEAT1 and NEAT2/MALAT1 exemplify long non-coding RNAs (lncRNAs) known to function in nuclear bodies; however, we suggest that RNA biogenesis itself may underpin much nuclear compartmentalization. Recent studies show that active genes cluster with nuclear speckles on a genome-wide scale, significantly advancing earlier cytological evidence that speckles (aka SC-35 domains) are hubs of concentrated pre-mRNA metabolism. We propose the 'karyotype to hub' hypothesis to explain this organization: clustering of genes in the human karyotype may have evolved to facilitate the formation of efficient nuclear hubs, driven in part by the propensity of ribonucleoproteins (RNPs) to form large-scale condensates. The special capacity of highly repetitive RNAs to impact architecture is highlighted by recent findings that human satellite II RNA sequesters factors into abnormal nuclear bodies in disease, potentially co-opting a normal developmental mechanism.
Keywords: Chromosome bands; Genome organization; Non-coding RNA; Nuclear structure; Speckles.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Conflict of interest statement Nothing declared.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

