Characterization of the AP2/ERF Transcription Factor Family and Expression Profiling of DREB Subfamily under Cold and Osmotic Stresses in Ammopiptanthus nanus
- PMID: 32260365
- PMCID: PMC7238250
- DOI: 10.3390/plants9040455
Characterization of the AP2/ERF Transcription Factor Family and Expression Profiling of DREB Subfamily under Cold and Osmotic Stresses in Ammopiptanthus nanus
Abstract
APETALA2/ethylene-responsive factor (AP2/ERF) is one of the largest transcription factor (TF) families in plants, which play important roles in regulating plant growth, development, and response to environmental stresses. Ammopiptanthus nanus, an unusual evergreen broad-leaved shrub in the arid region in the northern temperate zone, demonstrates a strong tolerance to low temperature and drought stresses, and AP2/ERF transcription factors may contribute to the stress tolerance of A. nanus. In the current study, 174 AP2/ERF family members were identified from the A. nanus genome, and they were divided into five subfamilies, including 92 ERF members, 55 dehydration-responsive element binding (DREB) members, 24 AP2 members, 2 RAV members, and 1 Soloist member. Compared with the other leguminous plants, A. nanus has more members of the DREB subfamily and the B1 group of the ERF subfamily, and gene expansion in the AP2/ERF family is primarily driven by tandem and segmental duplications. Promoter analysis showed that many stress-related cis-acting elements existed in promoter regions of the DREB genes, implying that MYB, ICE1, and WRKY transcription factors regulate the expression of DREB genes in A. nanus. Expression profiling revealed that the majority of DREB members were responsive to osmotic and cold stresses, and several DREB genes such as EVM0023336.1 and EVM0013392.1 were highly induced by cold stress, which may play important roles in cold response in A. nanus. This study provided important data for understanding the evolution and functions of AP2/ERF and DREB transcription factors in A. nanus.
Keywords: AP2/ERF; Ammopiptanthus nanus; cold stress; gene family; osmotic stress; transcription factor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

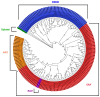

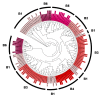


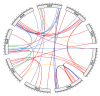
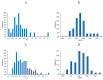


References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

