Adaptive Evolution of Geobacter sulfurreducens in Coculture with Pseudomonas aeruginosa
- PMID: 32265334
- PMCID: PMC7157779
- DOI: 10.1128/mBio.02875-19
Adaptive Evolution of Geobacter sulfurreducens in Coculture with Pseudomonas aeruginosa
Abstract
Interactions between microorganisms in mixed communities are highly complex, being either syntrophic, neutral, predatory, or competitive. Evolutionary changes can occur in the interaction dynamics between community members as they adapt to coexistence. Here, we report that the syntrophic interaction between Geobacter sulfurreducens and Pseudomonas aeruginosa coculture change in their dynamics over evolutionary time. Specifically, Geobacter sp. dominance increases with adaptation within the cocultures, as determined through quantitative PCR and fluorescence in situ hybridization. This suggests a transition from syntrophy to competition and demonstrates the rapid adaptive capacity of Geobacter spp. to dominate in cocultures with P. aeruginosa Early in coculture establishment, two single-nucleotide variants in the G. sulfurreducensfabI and tetR genes emerged that were strongly selected for throughout coculture evolution with P. aeruginosa phenazine wild-type and phenazine-deficient mutants. Sequential window acquisition of all theoretical spectra-mass spectrometry (SWATH-MS) proteomics revealed that the tetR variant cooccurred with the upregulation of an adenylate cyclase transporter, CyaE, and a resistance-nodulation-division (RND) efflux pump notably known for antibiotic efflux. To determine whether antibiotic production was driving the increased expression of the multidrug efflux pump, we tested Pseudomonas-derived phenazine-1-carboxylic acid (PHZ-1-CA) for its potential to inhibit Geobacter growth and drive selection of the tetR and fabI genetic variants. Despite its inhibitory properties, PHZ-1-CA did not drive variant selection, indicating that other antibiotics may drive overexpression of the efflux pump and CyaE or that a novel role exists for these proteins in the context of this interaction.IMPORTANCEGeobacter and Pseudomonas spp. cohabit many of the same environments, where Geobacter spp. often dominate. Both bacteria are capable of extracellular electron transfer (EET) and play important roles in biogeochemical cycling. Although they recently in 2017 were demonstrated to undergo direct interspecies electron transfer (DIET) with one another, the genetic evolution of this syntrophic interaction has not been examined. Here, we use whole-genome sequencing of the cocultures before and after adaptive evolution to determine whether genetic selection is occurring. We also probe their interaction on a temporal level and determine whether their interaction dynamics change over the course of adaptive evolution. This study brings to light the multifaceted nature of interactions between just two microorganisms within a controlled environment and will aid in improving metabolic models of microbial communities comprising these two bacteria.
Keywords: competition; evolution; mutualism; syntrophs.
Copyright © 2020 Semenec et al.
Figures
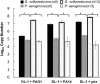
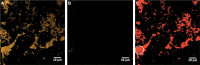
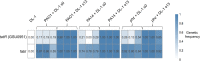
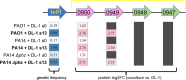
References
-
- Deng DD, Zhang YC, Liu Y. 2015. A Geobacter strain isolated from rice paddy soil with higher bioelectricity generation capability in comparison to Geobacter sulfurreducens PCA. RSC Adv 5:43978–43989. doi:10.1039/C5RA06211J. - DOI
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
