Fungal Dysbiosis and Intestinal Inflammation in Children With Beta-Cell Autoimmunity
- PMID: 32265922
- PMCID: PMC7103650
- DOI: 10.3389/fimmu.2020.00468
Fungal Dysbiosis and Intestinal Inflammation in Children With Beta-Cell Autoimmunity
Abstract
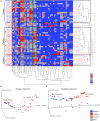
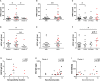
Although gut bacterial dysbiosis is recognized as a regulator of beta-cell autoimmunity, no data is available on fungal dysbiosis in the children at the risk of type 1 diabetes (T1D). We hypothesized that the co-occurrence of fungal and bacterial dysbiosis contributes to the intestinal inflammation and autoimmune destruction of insulin-producing beta-cells in T1D. Fecal and blood samples were collected from 26 children tested positive for at least one diabetes-associated autoantibody (IAA, GADA, IA-2A or ICA) and matched autoantibody-negative children with HLA-conferred susceptibility to T1D (matched for HLA-DQB1 haplotype, age, gender and early childhood nutrition). Bacterial 16S and fungal ITS2 sequencing, and analyses of the markers of intestinal inflammation, namely fecal human beta-defensin-2 (HBD2), calprotectin and secretory total IgA, were performed. Anti-Saccharomyces cerevisiae antibodies (ASCA) and circulating cytokines, IFNG, IL-17 and IL-22, were studied. After these analyses, the children were followed for development of clinical T1D (median 8 years and 8 months). Nine autoantibody positive children were diagnosed with T1D, whereas none of the autoantibody negative children developed T1D during the follow-up. Fungal dysbiosis, characterized by high abundance of fecal Saccharomyces and Candida, was found in the progressors, i.e., children with beta-cell autoimmunity who during the follow-up progressed to clinical T1D. These children showed also bacterial dysbiosis, i.e., increased Bacteroidales and Clostridiales ratio, which was, however, found also in the non-progressors, and is thus a common nominator in the children with beta-cell autoimmunity. Furthermore, the progressors showed markers of intestinal inflammation detected as increased levels of fecal HBD2 and ASCA IgG to fungal antigens. We conclude that the fungal and bacterial dysbiosis, and intestinal inflammation are associated with the development of T1D in children with beta-cell autoimmunity.
Keywords: Candida; Saccharomyces; dysbiosis; gut; inflammation; mycobiome; type 1 diabetes.
Copyright © 2020 Honkanen, Vuorela, Muthas, Orivuori, Luopajärvi, Tejesvi, Lavrinienko, Pirttilä, Fogarty, Härkönen, Ilonen, Ruohtula, Knip, Koskimäki and Vaarala.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

