Engineering of Trichoderma reesei for enhanced degradation of lignocellulosic biomass by truncation of the cellulase activator ACE3
- PMID: 32266008
- PMCID: PMC7110754
- DOI: 10.1186/s13068-020-01701-3
Engineering of Trichoderma reesei for enhanced degradation of lignocellulosic biomass by truncation of the cellulase activator ACE3
Abstract
Background: The filamentous fungus Trichoderma reesei is a major workhorse employed to produce cellulase, which hydrolyzes lignocellulosic biomass for the production of cellulosic ethanol and bio-based products. However, the economic efficiency of biorefineries is still low.
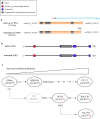
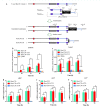
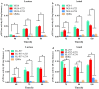
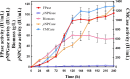
Results: In this study, the truncation of cellulase activator ACE3 was identified and characterized in T. reesei classical mutant NG14 and its direct descendants for the first time. We demonstrated that the truncated ACE3 is the crucial cause of cellulase hyper-production in T. reesei NG14 branch. Replacing the native ACE3 with truncated ACE3 in other T. reesei strains remarkably improves cellulase production. By truncating ACE3, we engineered a T. reesei mutant, PC-3-7-A723, capable of producing more cellulase than other strains. In a 30-L fermenter, fed-batch fermentation with PC-3-7-A723 drastically increased the maximum cellulase titer (FPase) to 102.63 IU/mL at 240 h, which constitutes a 20-30% improvement to that of the parental strain PC-3-7.
Conclusions: This work characterized the function of truncated ACE3 and demonstrated that analysis of classical mutants allows rational engineering of mutant strains with improved cellulase production necessary to process lignocellulosic biomass. Our rational engineering strategy might be useful for enhancing the production of other bio-based products.
Keywords: Cellulase production; Genetic engineering; Lignocellulosic biomass; Trichoderma reesei; Truncated ACE3.
© The Author(s) 2020.
Conflict of interest statement
Competing interestsThe authors declare no conflict of interest.
Figures

References
-
- de Paula RG, Antoniêto ACC, Ribeiro LFC, Srivastava N, O’Donovan A, Mishra PK, Gupta VK, Silva RN. Engineered microbial host selection for value-added bioproducts from lignocellulose. Biotechnol Adv. 2019;37:107347. - PubMed
-
- Yang X, Xu M, Yang S. Metabolic and process engineering of clostridium cellulovorans for biofuel production from cellulose. Metab Eng. 2015;32:39–48. - PubMed
-
- Kim SR, Skerker JM, Kong II, Kim H, Maurer MJ, Zhang GC, et al. Metabolic engineering of a haploid strain derived from a triploid industrial yeast for producing cellulosic ethanol. Metab Eng. 2017;40:176–185. - PubMed
-
- Li J, Lin L, Sun T, Xu J, Ji J, Liu Q, Tian C. Direct production of commodity chemicals from lignocellulose using Myceliophthora thermophile. Metab Eng. 2019;05:007. - PubMed
-
- Liu G, Qu Y. Engineering of filamentous fungi for efficient conversion of lignocellulose: tools, recent advances and prospects. Biotechnol Adv. 2018;37:519–529. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

