Clinical and genomic insights into circulating tumor DNA-based alterations across the spectrum of metastatic hormone-sensitive and castrate-resistant prostate cancer
- PMID: 32268276
- PMCID: PMC7186589
- DOI: 10.1016/j.ebiom.2020.102728
Clinical and genomic insights into circulating tumor DNA-based alterations across the spectrum of metastatic hormone-sensitive and castrate-resistant prostate cancer
Abstract
Background: Metastatic prostate cancer is a clonally heterogeneous disease state characterized by progressive somatic perturbations. The aim of this study was to identify cell free DNA- (cfDNA-) based alterations and their associations with outcomes in progressive metastatic prostate cancer.
Methods: In this longitudinal prospective cohort study plasma cfDNA/circulating tumor DNA (ctDNA) was analyzed before, during, and after androgen deprivation therapy (ADT) in 4 independent patient groups ranging from untreated metastatic hormone sensitive prostate cancer (mHSPC) to metastatic castrate resistant prostate cancer (mCRPC). Next generation sequencing was performed on ctDNA and germline DNA to characterize alterations and associations with clinical outcomes were determined for each group.
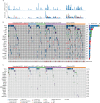
Findings: cfDNA yields were different in progressive mHSPC and mCRPC states (P < .001). In mHSPC, a higher than median ctDNA fraction was predictive of shorter time to ADT failure (HR, 2.29 [95% CI, 1.13-4.65]; Log-Rank P = .02). cfDNA, ctDNA taken with volume of metastatic disease in mHSPC and with alkaline phosphatase levels prognosticated survival better than clinical factors alone in mHSPC and mCRPC states (Log Rank P = 0.03). ctDNA-based AR, APC mutations were increased in mCRPC compared to mHSPC (P < ·05).TP53 mutations, RB1 loss, and AR gene amplifications correlated with poorer survival in mCRPC. Mutations in multiple DNA repair genes (ATM, BRCA1, BRCA2, CHEK2) were associated with time to ADT treatment failure and survival in mHSPC.
Interpretation: ctDNA fraction can further refine clinical prognostic factors in metastatic prostate cancer. Somatic ctDNA alterations have potential prognostic, predictive, and therapeutic implications in metastatic prostate cancer management.
Funding: Several funding sources have supported this study. A full list is provided in the Acknowledgments. No funding was received from Predicine, Inc. during the conduct of the study.
Keywords: Circulating tumor DNA; Genomic alterations; Metastatic prostate cancer.
Copyright © 2020 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest Manish Kohli received travel/accommodation from Celgene; Tiantian Zheng, Amy Wang, Carlos Montesinos, Calven Wong, Pan Du, Shidong Jia, and Jianjun Yu are stockholders in Predicine, Inc.; Lisa Horvath received research funding from Astellas Pharma, travel/accommodation from Astellas Pharma and Pfizer, and is on the Scientific Advisory Board for Imagion; Kate Mahon received travel/accommodation from Astellas Pharma; Edmond M Kwan received honoraria from Janssen, research funding from Astellas Pharma and AstraZeneca, and travel /accommodations from Astellas Pharma, Pfizer, and Ipsen; Arun A Azad is a consultant for Astellas Pharma, Janssen, and Novartis, is on the speakers bureau for Astellas Pharma, Janssen, Novartis and Amgen received honoraria from Astellas Pharma, Janssen, Novartis, Tolmar, Amgen, Pfizer, and Telix, is on the Scientific Advisory Board for Astellas Pharma, Novartis, Sanofi, AstraZeneca, Tolmar, Pfizer, and Telix, and received research funding from Astellas Pharma, and Merck Serono.
Figures




References
-
- Gandara D.R., Paul S.M., Kowanetz M., Schleifman E., Zou W., Li Y. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat Med. 2018;24(9):1441–1448. - PubMed
-
- Gandara D.R., Paul S.M., Kowanetz M., Schleifman E., Zou W., Li Y. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat. Med. 2018;24(9):1441. - PubMed
-
- Basch E., Loblaw D.A., Rumble R.B. Systemic therapy in men with metastatic castration-resistant prostate cancer: American Society of Clinical Oncology and Cancer Care Ontario Clinical Practice guideline summary. J Oncol Pract. 2014;10(6):e418–ee20. - PubMed
-
- Davis I.D., Martin A.J., Stockler M.R., Begbie S., Chi K.N., Chowdhury S. Enzalutamide with standard first-line therapy in metastatic prostate cancer. New Engl J Med. 2019;381(2):121–131. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

