Generation of synthetic nanobodies against delicate proteins
- PMID: 32269381
- PMCID: PMC7617899
- DOI: 10.1038/s41596-020-0304-x
Generation of synthetic nanobodies against delicate proteins
Abstract
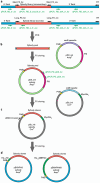
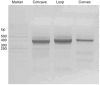
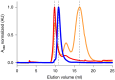
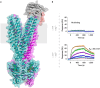
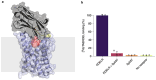
Here, we provide a protocol to generate synthetic nanobodies, known as sybodies, against any purified protein or protein complex within a 3-week period. Unlike methods that require animals for antibody generation, sybody selections are carried out entirely in vitro under controlled experimental conditions. This is particularly relevant for the generation of conformation-specific binders against labile membrane proteins or protein complexes and allows selections in the presence of non-covalent ligands. Sybodies are especially suited for cases where binder generation via immune libraries fails due to high sequence conservation, toxicity or insufficient stability of the target protein. The procedure entails a single round of ribosome display using the sybody libraries encoded by mRNA, followed by two rounds of phage display and binder identification by ELISA. The protocol is optimized to avoid undesired reduction in binder diversity and enrichment of non-specific binders to ensure the best possible selection outcome. Using the efficient fragment exchange (FX) cloning method, the sybody sequences are transferred from the phagemid to different expression vectors without the need to amplify them by PCR, which avoids unintentional shuffling of complementary determining regions. Using quantitative PCR (qPCR), the efficiency of each selection round is monitored to provide immediate feedback and guide troubleshooting. Our protocol can be carried out by any trained biochemist or molecular biologist using commercially available reagents and typically gives rise to 10-30 unique sybodies exhibiting binding affinities in the range of 500 pM-500 nM.
Conflict of interest statement
The authors declare competing financial interests. I.Z., P.E., R.J.P.D. and M.A.S. are co-founders and shareholders of Linkster Therapeutics AG.
Figures
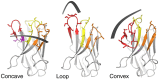

References
-
- Ries J, Kaplan C, Platonova E, Eghlidi H, Ewers H. A simple, versatile method for GFP-based super-resolution microscopy via nanobodies. Nat Methods. 2012;9:582–584. - PubMed
-
- Rothbauer U, et al. Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat Methods. 2006;3:887–889. - PubMed
-
- Bukowska MA, Grutter MG. New concepts and aids to facilitate crystallization. Curr Opin Struct Biol. 2013;23:409–416. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

