Prevalence of Antibiotic Resistome in Ready-to-Eat Salad
- PMID: 32269985
- PMCID: PMC7109403
- DOI: 10.3389/fpubh.2020.00092
Prevalence of Antibiotic Resistome in Ready-to-Eat Salad
Abstract
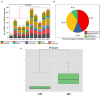
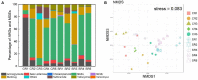
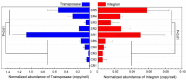
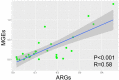
Ready-to-eat salad harbors microorganisms that may carry various antibiotic resistance genes (ARGs). However, few studies have focused on the prevalence of ARGs on salad, thus underestimating the risk of ARGs transferring from salad to consumers. In this small-scale study, high-throughput quantitative PCR was used to explore the presence, prevalence and abundance of ARGs associated with serving salad sourced from two restaurant types, fast-food chain and independent casual dining. A total of 156 unique ARGs and nine mobile genetic elements (MGEs) were detected on the salad items assessed. The abundance of ARGs and MGEs were significantly higher in independent casual dining than fast-food chain restaurants. Absolute copies of ARGs in salad were 1.34 × 107 to 2.71 × 108 and 1.90 × 108 to 4.87 × 108 copies per g salad in fast-food and casual dining restaurants, respectively. Proteobacteria, Bacteroidetes, Actinobacteria, and Firmicutes were the dominant bacterial phyla detected from salad samples. Pseudomonas, Acinetobacter, Exiguobacterium, Weissella, Enterobacter, Leuconostoc, Pantoea, Serratia, Erwinia, and Ewingella were the 10 most dominant bacterial genera found in salad samples. A significant positive correlation between ARGs and MGEs was detected. These results integrate knowledge about the ARGs in ready-to-eat salad and highlight the potential impact of ARGs transfer to consumers.
Keywords: antibiotic resistance genes (ARGs); high-throughput quantitative PCR; human health; ready-to-eat food; salad.
Copyright © 2020 Zhou, Wei, Giles, Neilson, Zheng, Zhang, Zhu and Yang.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

