Virus-host coexistence in phytoplankton through the genomic lens
- PMID: 32270031
- PMCID: PMC7112755
- DOI: 10.1126/sciadv.aay2587
Virus-host coexistence in phytoplankton through the genomic lens
Abstract
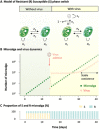
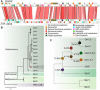
Virus-microbe interactions in the ocean are commonly described by "boom and bust" dynamics, whereby a numerically dominant microorganism is lysed and replaced by a virus-resistant one. Here, we isolated a microalga strain and its infective dsDNA virus whose dynamics are characterized instead by parallel growth of both the microalga and the virus. Experimental evolution of clonal lines revealed that this viral production originates from the lysis of a minority of virus-susceptible cells, which are regenerated from resistant cells. Whole-genome sequencing demonstrated that this resistant-susceptible switch involved a large deletion on one chromosome. Mathematical modeling explained how the switch maintains stable microalga-virus population dynamics consistent with their observed growth pattern. Comparative genomics confirmed an ancient origin of this "accordion" chromosome despite a lack of sequence conservation. Together, our results show how dynamic genomic rearrangements may account for a previously overlooked coexistence mechanism in microalgae-virus interactions.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures



References
-
- Suttle C. A., Marine viruses—Major players in the global ecosystem. Nat. Rev. Microbiol. 5, 801–812 (2007). - PubMed
-
- Brussaard C. P. D., Wilhelm S. W., Thingstad F., Weinbauer M. G., Bratbak G., Heldal M., Kimmance S. A., Middelboe M., Nagasaki K., Paul J. H., Schroeder D. C., Suttle C. A., Vaqué D., Wommack K. E., Global-scale processes with a nanoscale drive: The role of marine viruses. ISME J. 2, 575–578 (2008). - PubMed
-
- Hingamp P., Grimsley N., Acinas S. G., Clerissi C., Subirana L., Poulain J., Ferrera I., Sarmento H., Villar E., Lima-Mendez G., Faust K., Sunagawa S., Claverie J.-M., Moreau H., Desdevises Y., Bork P., Raes J., de Vargas C., Karsenti E., Kandels-Lewis S., Jaillon O., Not F., Pesant S., Wincker P., Ogata H., Exploring nucleo-cytoplasmic large DNA viruses in Tara Oceans microbial metagenomes. ISME J. 7, 1678–1695 (2013). - PMC - PubMed
-
- Zhu F., Massana R., Not F., Marie D., Vaulot D., Mapping of picoeucaryotes in marine ecosystems with quantitative PCR of the 18S rRNA gene. FEMS Microbiol. Ecol. 52, 79–92 (2005). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

