Blind estimation and correction of microarray batch effect
- PMID: 32271844
- PMCID: PMC7145015
- DOI: 10.1371/journal.pone.0231446
Blind estimation and correction of microarray batch effect
Abstract
Microarray batch effect (BE) has been the primary bottleneck for large-scale integration of data from multiple experiments. Current BE correction methods either need known batch identities (ComBat) or have the potential to overcorrect, by removing true but unknown biological differences (Surrogate Variable Analysis SVA). It is well known that experimental conditions such as array or reagent batches, PCR amplification or ozone levels can affect the measured expression levels; often the direction of perturbation of the measured expression is the same in different datasets. However, there are no BE correction algorithms that attempt to estimate the individual effects of technical differences and use them to correct expression data. In this manuscript, we show that a set of signatures, each of which is a vector the length of the number of probes, calculated on a reference set of microarray samples can predict much of the batch effect in other validation sets. We present a rationale of selecting a reference set of samples designed to estimate technical differences without removing biological differences. Putting both together, we introduce the Batch Effect Signature Correction (BESC) algorithm that uses the BES calculated on the reference set to efficiently predict and remove BE. Using two independent validation sets, we show that BESC is capable of removing batch effect without removing unknown but true biological differences. Much of the variations due to batch effect is shared between different microarray datasets. That shared information can be used to predict signatures (i.e. directions of perturbation) due to batch effect in new datasets. The correction can be precomputed without using the samples to be corrected (blind), done on each sample individually (single sample) and corrects only known technical effects without removing known or unknown biological differences (conservative). Those three characteristics make it ideal for high-throughput correction of samples for a microarray data repository. We also compare the performance of BESC to three other batch correction methods: SVA, Removing Unwanted Variation (RUV) and Hidden Covariates with Prior (HCP). An R Package besc implementing the algorithm is available from http://explainbio.com.
Conflict of interest statement
The author (SV) does contract statistical analysis under the business name of “HiThru Analytics LLC”. Currently he is working as a contractor, part time with the National Institutes of Health (Bethesda MD) and part time with Tridiuum Inc. (Philadelphia PA). This does not alter our adherence to PLOS ONE policies on sharing data and materials.
Figures
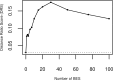

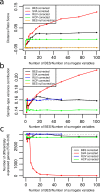
Similar articles
-
Preserving biological heterogeneity with a permuted surrogate variable analysis for genomics batch correction.Bioinformatics. 2014 Oct;30(19):2757-63. doi: 10.1093/bioinformatics/btu375. Epub 2014 Jun 6. Bioinformatics. 2014. PMID: 24907368 Free PMC article.
-
Removing batch effects from purified plasma cell gene expression microarrays with modified ComBat.BMC Bioinformatics. 2015 Feb 25;16:63. doi: 10.1186/s12859-015-0478-3. BMC Bioinformatics. 2015. PMID: 25887219 Free PMC article.
-
Batch effect reduction of microarray data with dependent samples using an empirical Bayes approach (BRIDGE).Stat Appl Genet Mol Biol. 2021 Dec 14;20(4-6):101-119. doi: 10.1515/sagmb-2021-0020. Stat Appl Genet Mol Biol. 2021. PMID: 34905304 Free PMC article.
-
Why Batch Effects Matter in Omics Data, and How to Avoid Them.Trends Biotechnol. 2017 Jun;35(6):498-507. doi: 10.1016/j.tibtech.2017.02.012. Epub 2017 Mar 25. Trends Biotechnol. 2017. PMID: 28351613 Review.
-
Perspectives for better batch effect correction in mass-spectrometry-based proteomics.Comput Struct Biotechnol J. 2022 Aug 12;20:4369-4375. doi: 10.1016/j.csbj.2022.08.022. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36051874 Free PMC article. Review.
Cited by
-
Differential expression of ion channel coding genes in the endometrium of women experiencing recurrent implantation failures.Sci Rep. 2024 Aug 27;14(1):19822. doi: 10.1038/s41598-024-70778-9. Sci Rep. 2024. PMID: 39192025 Free PMC article.
-
Comprehensive characterization of adipogenesis-related genes in colorectal cancer for clinical significance and immunogenomic landscape analyses.Lipids Health Dis. 2023 Dec 7;22(1):217. doi: 10.1186/s12944-023-01942-9. Lipids Health Dis. 2023. PMID: 38062499 Free PMC article.
-
High expression of collagen 1A2 promotes the proliferation and metastasis of esophageal cancer cells.Ann Transl Med. 2020 Dec;8(24):1672. doi: 10.21037/atm-20-7867. Ann Transl Med. 2020. PMID: 33490184 Free PMC article.
-
Assessing and mitigating batch effects in large-scale omics studies.Genome Biol. 2024 Oct 3;25(1):254. doi: 10.1186/s13059-024-03401-9. Genome Biol. 2024. PMID: 39363244 Free PMC article. Review.
-
Novel ribosome biogenesis-related biomarkers and therapeutic targets identified in psoriasis.Sci Rep. 2025 May 27;15(1):18525. doi: 10.1038/s41598-025-03833-8. Sci Rep. 2025. PMID: 40425712 Free PMC article.
References
-
- Scherer A. Batch Effects and Noise in Microarray Experiments: Sources and Solutions. http://www.wiley.com/WileyCDA/WileyTitle/productCd-0470741384.html. Accessed 29 Nov 2016.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

