Complete Mitogenomic Structure and Phylogenetic Implications of the Genus Ostrinia (Lepidoptera: Crambidae)
- PMID: 32272743
- PMCID: PMC7240680
- DOI: 10.3390/insects11040232
Complete Mitogenomic Structure and Phylogenetic Implications of the Genus Ostrinia (Lepidoptera: Crambidae)
Abstract
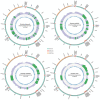
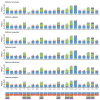
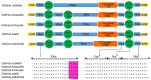
To understand mitogenome characteristics and reveal phylogenetic relationships of the genus Ostrinia, including several notorious pests of great importance for crops, we sequenced the complete mitogenomes of four species: Ostrinia furnacalis (Guenée, 1854), Ostrinia nubilalis (Hübner, 1796), Ostrinia scapulalis (Walker, 1859) and Ostrinia zealis (Guenée, 1854). Results indicate that the four mitogenomes-O. furnacalis, O. nubilalis, O. scapulalis, and O. zealis-are 15,245, 15,248, 15,311, and 15,208 bp in size, respectively. All four mitogenomes are comprised of 37 encoded genes and a control region. All 13 protein-coding genes (PCGs) initiate with ATN and terminate with TAN, with the exception of cox1 that starts with CGA, and cox1, cox2, and nad5 that terminate with an incomplete codon T. All transfer RNA genes (tRNAs) present the typical clover-leaf secondary structure except for the trnS1 (AGN) gene. There are some conserved structural elements in the control region. Our analyses indicate that nad6 and atp6 exhibit higher evolution rates compared to other PCGs. Phylogenetic analyses based on mitogenomes using both maximum likelihood (ML) and Bayesian inference (BI) methods revealed the relationship (O. palustralis + (O. penitalis + (O. zealis + (O. furnacalis + (O. nubilalis + O. scapulalis))))) within Ostrinia.
Keywords: Crambidae; Ostrinia; mitochondrial genome; phylogenetic analysis.
Conflict of interest statement
All authors declare no conflicting interests.
Figures



Similar articles
-
Complete mitochondrial genome of Ostrinia kasmirica (Lepidoptera: Crambidae).Mitochondrial DNA B Resour. 2021 Jul 9;6(8):2316-2318. doi: 10.1080/23802359.2021.1950058. eCollection 2021. Mitochondrial DNA B Resour. 2021. PMID: 34291169 Free PMC article.
-
Characterization of the complete mitochondrial genome of Asia Corn Borer, Ostrinia furnacalis (Lepidoptera: Crambidae).Mitochondrial DNA B Resour. 2020 Jan 31;5(1):936-937. doi: 10.1080/23802359.2020.1718025. Mitochondrial DNA B Resour. 2020. PMID: 33366817 Free PMC article.
-
Characterization of four mitochondrial genomes of Crambidae (Lepidoptera, Pyraloidea) and phylogenetic implications.Arch Insect Biochem Physiol. 2023 Jan;112(1):e21914. doi: 10.1002/arch.21914. Epub 2022 May 15. Arch Insect Biochem Physiol. 2023. PMID: 35570199
-
Complete mitochondrial genome of Phyllonorycter ringoniella (Lepidoptera: Gracillariidae).Mitochondrial DNA B Resour. 2022 May 10;7(5):798-800. doi: 10.1080/23802359.2022.2072245. eCollection 2022. Mitochondrial DNA B Resour. 2022. PMID: 35573596 Free PMC article.
-
Characterization of the complete mitochondrial genome of Tryporyza incertulas, in comparison with seven other Pyraloidea moths.Gene. 2014 Jan 1;533(1):356-65. doi: 10.1016/j.gene.2013.07.072. Epub 2013 Aug 15. Gene. 2014. PMID: 23954873
Cited by
-
Novel insight into lepidopteran phylogenetics from the mitochondrial genome of the apple fruit moth of the family Argyresthiidae.BMC Genomics. 2024 Jan 2;25(1):21. doi: 10.1186/s12864-023-09905-1. BMC Genomics. 2024. PMID: 38166583 Free PMC article.
-
Comparative Analysis of Mitochondrial Genomes among Twelve Sibling Species of the Genus Atkinsoniella Distant, 1908 (Hemiptera: Cicadellidae: Cicadellinae) and Phylogenetic Analysis.Insects. 2022 Mar 3;13(3):254. doi: 10.3390/insects13030254. Insects. 2022. PMID: 35323552 Free PMC article.
-
The complete mitogenome of Curculiochinensis (Chevrolat, 1878) (Coleoptera: Curculionidae: Curculioninae).Biodivers Data J. 2021 Oct 25;9:e69196. doi: 10.3897/BDJ.9.e69196. eCollection 2021. Biodivers Data J. 2021. PMID: 34759727 Free PMC article.
-
Complete mitochondrial genome of Ostrinia kasmirica (Lepidoptera: Crambidae).Mitochondrial DNA B Resour. 2021 Jul 9;6(8):2316-2318. doi: 10.1080/23802359.2021.1950058. eCollection 2021. Mitochondrial DNA B Resour. 2021. PMID: 34291169 Free PMC article.
-
Evolution of the Sex Pheromone Communication System in Ostrinia Moths.Insects. 2021 Nov 28;12(12):1067. doi: 10.3390/insects12121067. Insects. 2021. PMID: 34940155 Free PMC article. Review.
References
-
- Yang R.S., Wang Z.Y., He K.L. Advances in phylogenetic and taxonomic studies on genus Ostrinia. Plant Prot. Beijing. 2007;33:20–25. doi: 10.3969/j.issn.0529-1542.2007.02.005. - DOI
-
- Mutuura A., Munroe E. Taxonomy and distribution of the European corn borer and allied species: Genus Ostrinia (Lepidoptera: Pyralidae) Mem. Entomol. Soc. Can. 1970;102:1–112. doi: 10.4039/entm10271fv. - DOI
-
- Park K.T. Taxonomic study of the corn stem borer in Korea with allied species of the genus Ostrinia (Lep.; Pyralidae) Korean. J. Appl. Entomol. 1975;14:221–225.
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

