Comparative analysis of targeted next-generation sequencing panels for the detection of gene mutations in chronic lymphocytic leukemia: an ERIC multi-center study
- PMID: 32273480
- PMCID: PMC7927885
- DOI: 10.3324/haematol.2019.234716
Comparative analysis of targeted next-generation sequencing panels for the detection of gene mutations in chronic lymphocytic leukemia: an ERIC multi-center study
Abstract
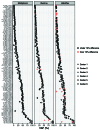
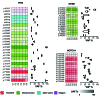
Next-generation sequencing (NGS) has transitioned from research to clinical routine, yet the comparability of different technologies for mutation profiling remains an open question. We performed a European multicenter (n=6) evaluation of three amplicon-based NGS assays targeting 11 genes recurrently mutated in chronic lymphocytic leukemia. Each assay was assessed by two centers using 48 pre-characterized chronic lymphocytic leukemia samples; libraries were sequenced on the Illumina MiSeq instrument and bioinformatics analyses were centralized. Across all centers the median percentage of target reads ≥100x ranged from 94.2- 99.8%. In order to rule out assay-specific technical variability, we first assessed variant calling at the individual assay level i.e., pairwise analysis of variants detected amongst partner centers. After filtering for variants present in the paired normal sample and removal of PCR/sequencing artefacts, the panels achieved 96.2% (Multiplicom), 97.7% (TruSeq) and 90% (HaloPlex) concordance at a variant allele frequency (VAF) >0.5%. Reproducibility was assessed by looking at the inter-laboratory variation in detecting mutations and 107 of 115 (93% concordance) mutations were detected by all six centers, while the remaining eight variants (7%) were undetected by a single center. Notably, 6 of 8 of these variants concerned minor subclonal mutations (VAF <5%). We sought to investigate low-frequency mutations further by using a high-sensitivity assay containing unique molecular identifiers, which confirmed the presence of several minor subclonal mutations. Thus, while amplicon-based approaches can be adopted for somatic mutation detection with VAF >5%, after rigorous validation, the use of unique molecular identifiers may be necessary to reach a higher sensitivity and ensure consistent and accurate detection of low-frequency variants.
Figures

Comment in
-
Validating genomic tools for precision medicine in chronic lymphocytic leukemia: ERIC leads the way.Haematologica. 2021 Mar 1;106(3):656-658. doi: 10.3324/haematol.2020.270652. Haematologica. 2021. PMID: 33645943 Free PMC article. No abstract available.
References
-
- Quesada V, Conde L, Villamor N, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet. 2011;44(1):47-52. - PubMed
-
- Puente XS, Bea S, Valdes-Mas R, et al. Noncoding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015; 526(7574):519-524. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

